Original Articles: 2023 Vol: 15 Issue: 1
Virtual Screening, ADMET Analysis and Synthesis of 2-(1H- benzotriazol-1-yl) N- Substituted Acetohydrazide that Bind to the Glycoprotein B of Herpes Simplex Virus-I (HSV-I)
Mali Dhanashri R*, Amrutkar Sunil V
Department of Pharmaceutical Chemistry, Gokhale Education Society’s Sir Dr. M. S. Gosavi College of Pharmaceutical Education and Research, Nashik, India
- Corresponding Author:
- Mali Dhanashri R
Department of Pharmaceutical Chemistry,
Gokhale Education Society’s Sir Dr. M. S. Gosavi College of Pharmaceutical Education and Research,
Nashik,
India
Received: 03-Jan-2023, Manuscript No. JOCPR-22-82379; Editor assigned: 06-Jan-2023, JOCPR-22-82379 (PQ); Reviewed: 13-Jan-2023, QC No. JOCPR-22-82379; Revised: 27-Jan-2023, Manuscript No. JOCPR-22-82379 (R); Published: 10-Feb-2023
Abstract
Background: Herpes Simplex Virus type 1 (HSV-1) is a contagious human pathogen causing serious infection. In recent decade the virus become dormant and resistant to available treatment, creates the need for development of new molecules. Benzotriazole is versatile molecule with wide range of activities like antibacterial, antiprotozoal, antiulcer, anthelmintic and antiproliferative activity. Result: A series of 2-(1H- benzotriazol-1-yl) N-substituted acetohydrazide derivatives were synthesized in good yield and were characterized by spectral methods and elemental analysis. The antiviral activity against the glycoprotein B of Herpes Simplex Virus-I (HSV-I) was determined by using molecular docking (2GUM). All compounds are having strong binding affinity over the standard acyclovir. Compound 5 h has highest binding affinity and have highest inhibitory activity. Conclusion: The benzotriazole-N substituted acetohydrazide derivatives has highest binding affinity towards glycoprotein B of Herpes Simplex Virus-I (HSV-I), stronger inhibition that makes a good antiviral agent.
Keywords
Benzotriazole; Herpes Simplex Virus-I (HSV-I); Antiviral; In silico activity; Molecular docking
Introduction
Herpes virus is a double stranded DNA virus. It belongs to family herpesviridae which has three different subfamilies namely the alpha, beta and gamma herpes viruses. HSV-1 is DNA virus with an icosahedral capsid, a tegument and an envelope. The envelope is made up of viral glycoproteins and host membrane components. Herpes Simplex Virus type 1 (HSV-1) is a contagious human pathogen that is estimated to affect 3.7 billion people worldwide [1-3].
Although the majority of infections are asymptomatic, HSV-1 infections afflict a significant portion of the human population and invade epithelial cells, causing cold sores or lesions on the eyelid. HSV-1 can also result in keratitis, which can cause blindness or severe visual impairment. Less commonly, encephalitis caused by CNS infection is linked to HSV-1.
Whereas HSV-1 infections are commonly associated with limited facial oral lesions, severe disease can occur in neonates or immunocompromised individuals (keratitis, meningitis, encephalitis and disseminated infections [4-6].
In recent years, shortage of specific antivirals for various viral infections clearly indicate the space and scope for future research in novel antiviral drug discovery as the scientific society is struggling and unable to battle against the viruses.
Nucleoside derivatives including acyclovir, ganciclovir, penciclovir, valaciclovir and famciclovir was used to treat Herpes Simplex Virus (HSV) infection [7-9].
Herpes Simplex Virus 1 (HSV-1) is a double stranded DNA virus with an icosahedral capsid, covered by the tegument and envelope, the latter of which is composed of host membrane components and viral glycoproteins.
MATERIALS AND METHODS
All reagents and solvents were used without further purification. All the recorded melting points were taken in an open glass capillary on a Griffin apparatus and the values given were uncorrected [10].
IR spectra were determined using potassium bromide discs and values were represented in cm-1. IR spectra were recorded on Shimadzu IR 435 spectrophotometer. 1H NMR spectra were carried out on Bruker 400 MHz spectrophotometer. Chemical shifts were recorded in ppm on δ-scale. The reaction progress was monitored by TLC using precoated silica gel aluminium sheets (MERCK). The spots were visualized using UV lamp. The solvent system used was ethanol and ethyl acetate and with a ratio of 2:3.
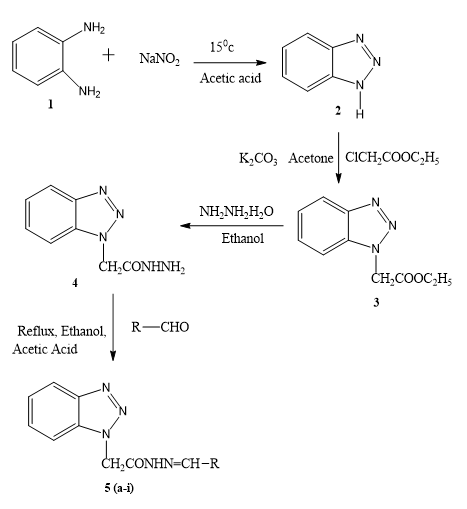
Synthesis of 1H-1,2,3-benzotriazole (2)
OPD (1) (0.1 mole) was dissolved in a mixture of glacial acetic acid (0.2 mol) and 30 ml of water. Clear solution was cooled to 15°C and then it was stirred magnetically and added to a solution of sodium nitrate (0.11 mol) in 15 ml of water. Reaction mixture became warmed and within 2 mins-3 mins reached a temperature of about 85°C, cooling was started with the change of colour from deep red to pale brown. Stirring was continued for 15 min and then it was chilled in ice water bath for 30 min. Pale brown precipitate of benzotriazole (1) separates out which was washed with ice cold water. Recrystallized using boiling water, white shiny needle shape crystal, 67.90%, m.p. 96°C-99°C, Rf 0.56 [11-13].
Synthesis of ethyl-1H-benzotriazol-1-yl acetate (3)
A mixture of 1H benzotriazole (2) (0.05 mol), chloroethyl acetate (0.05 mol) and potassium carbonate (1g) was stirred in dry acetone for 6 hrs. It was extracted with ether and solvent removed under pressure to obtain needle shaped white crystal. White shiny needle shape crystal, 72%, m.p. 40°C-44°C Rf 0.67.
Synthesis of 2-(1H- benzotriazol-1-yl) acetohydrazide (4)
Compound 3 (0.05 mol) and hydrazine hydrate (0.15 mol) in ethanol was stirred for 1 hr. and then refluxed for 3 hr. Then excessive solvent removed under pressure and it was recrystallized by mixture of chloroform hexane. White shiny crystal, 67%, m.p.130°C-132°C, Rf 0.78.
Synthesis of 2-(1H-benzotriazol-1-yl) N-substituted acetohydrazide (5 a-i)
Equimolar quantities of substituted aldehyde and compound 4 were dissolved in warm ethanol containing 0.5 ml of glacial acetic acid. The mixture was refluxed for 6 hr. and set aside. The resultant solid was recrystallized from ethanol (Figure 1).
In silico ADMET screening
The ADMET analysis was carried out in order to check for the nature and possible toxicity of the synthesized derivatives while administering in vivo. Online chemical property calculator mol inspirations were used to asses physicochemical properties and other pharmacokinetic properties and toxicity were assed using the PreADMET server and Swiss ADME [14].
Molecular docking
3D structure of tyrosinase was downloaded from the PDB site (PDB entry 3NM8).
Retrieval of target protein molecule
The 3D structure of glycoprotein B of herpes simplex virus- I (PDB ID: 2GUM) has been acquired from protein data bank this protein is a homotrimer (Chain A, B and C) with 628 amino acid residues and one sodium ion as an attached ligand. X-Ray Diffraction was used to determine the structure at a resolution of 2.10Å.
Active site prediction
Pymol software was used to invent the active site of the target protein and discovery studio visualizer was used find the active site amino acid that was used for creating the grid box. The active site of glycoprotein reside in domain II consisting of chain C.
Target protein preparation
Water molecule, bonded ligand and heteroatoms are removed from protein molecule in discovery studio visualizer as the molecule saved in pdb format.
Screening and retrieval of ligands
2D structures of synthesized derivatives were drawn using Chem draw ultra-program V.12.0.2 and then saved in sdf format. Then it converted to the docking compatible formats PDB using Open Babel (version 3.0.0). In order to prepare the ligands, the charged group of the ligands was neutralized and hydrogen was then added. Finally, the prepared ligands were docked using AutoDock 4.2 [15-17].
Molecular docking protocol
A ligand molecule library was screened against glycoprotein B (2GUM) using AutoDock 4.2. For identifying the interaction of ligand molecules with targeted proteins, the searching grid for the active site was built according to the visualization in discovery studio. The grid box was centered at the binding site and was 40 Å × 40 Å × 40 Å. Then Kollman charges, Gasteiger type polar hydrogen charges and polar hydrogen were allocated along with internal degrees of freedom and torsions to the protein. In order to find the optimal orientation for a protein ligand interaction, rigid docking was then carried out using the Lamarckian genetic algorithm, with 10 runs. The results were evaluated by ranking the different complexes according to their predicted binding energy. A more in depth study of the protein ligand interaction was performed using the discovery studio visualizer tool. LIGPLOT was used to generate 2D image of the interaction.
RESULTS AND DISCUSSION
Chemistry
Benzotriazole which prepared by diazotization and condensation of o-Phenylenediamine with sodium nitrate. Then it is combined with hydrazine hydrate and finally the targeted molecule obtained by reacting compound 4 with various aldehyde in presence of acetic acid. Table 1 summarizes the physical properties of 2-(1H- benzotriazol-1-yl) N- substituted acetohydrazide derivatives. All of the synthesized compounds were obtained in high purity and yield.
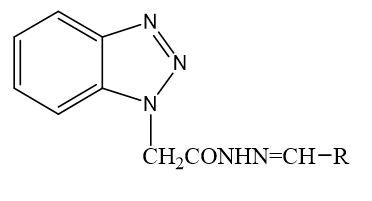
2-(1H-benzotriazol-1-yl) N-substituted acetohydrazide
| Comp Code | R | IUPAC name | Mp (°C) |
Yield (%) | Rf value |
|---|---|---|---|---|---|
| 5a | 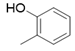 |
2(1H-benzotriazol-1-yl)-N’-(E)-(2-hydroxyphenyl) methylidene acetohydrazide. | 134-38 | 67 | 0.78 |
| 5b | 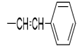 |
2(1H-benzotriazol-1-yl)-N’-(E)-ethylidene acetohydrazide. | 186-90 | 72 | 0.82 |
| 5c | 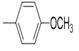 |
2(1H-benzotriazol-1-yl)-N’-(E)-(2-meth oxyphenyl) methylidene acetohydrazide. | 182-86 | 68 | 0.79 |
| 5d | 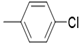 |
2(1H-benzotriazol-1-yl)-N’-(E)-(4- chloro) methylidene acetohydrazide. | 194-98 | 62 | 0.67 |
| 5e | 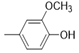 |
2(1H-benzotriazol-1-yl)-N’-(E)-(4-hydroxy-3- methoxyphenyl) methylidene acetohydrazide. | 170-74 | 65 | 0.71 |
| 5f | 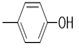 |
2(1H-benzotriazol-1-yl)-N’-(E)-(4-hydroxyphenyl) methylidene acetohydrazide. | 162-64 | 59 | 0.76 |
| 5g | 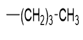 |
2(1H-benzotriazol-1-yl)-N’-(1E)-(pentylidene) methylidene acetohydrazide. | 174-78 | 63 | 0.59 |
| 5h | 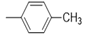 |
2(1H-benzotriazol-1-yl)-N’-(E)-(4-methylphenyl) methylidene acetohydrazide. | 210-12 | 66 | 0.73 |
| 5i | 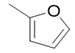 |
2(1H-benzotriazol-1-yl)-N’-(E)-(2-furan-2-yl) methylidene acetohydrazide. | 178-80 | 78 | 0.63 |
Table 1: Physical properties of synthesized 2-(1H-benzotriazol-1-yl) N-substituted acetohydrazide derivatives.
2(1H-benzotriazol-1-yl)-N’-(E)-(2-hydroxyphenyl) methylidene acetohydrazide (5a): 1813 (C=O amide str), 3317 (O-H Str), 3201 (N-H iminestr), 2785 (C-H str), 1620 (N=C iminestr, 1226 (C-O str), 1373 (C-N Str), 1HNMR (400 MHz, DMSO-d6) δ 10.82 (1H, OH), 10.56 (1H, NH), 8.76 (2H, NCH2), 7.86-8.02 (4H, Ar-H, Btz), 6.93-7.32 (4H, Ar-H), m/z 296.11 [18].
2(1H-benzotriazol-1-yl)-N’-(E)-ethylidene Acetohydrazide (5b): 1828 (C=O amide str), 3039 (N-H iminestr), 2800 (C-H str), 1627 (N=C imineStr, 1288 (C-O str), 1350(C-N Str), 3039(HC=CH Str).
2(1H-benzotriazol-1-yl)-N’-(E)-(2-meth oxyphenyl) methylidene Acetohydrazide (5C): 1890 (C=O amide str), 3194 (N-H iminestr), 2839 (C-H str), 1604 (N=C imineStr, 1257 (C-O str), 1311 (C-N Str), 1257(C-O-C str), 1HNMR (400 MHz, DMSO-d6) δ 10.91(1H, NH), 8.41(2H, NCH2), 7.56-8.22(4H, Ar-H, Btz), 7.07-7.72(4H, Ar-H), 5.60(1H, CH2), 3.45 (3H, OCH3), m/z 310.13 [19].
2(1H-benzotriazol-1-yl)-N’-(E)-(4-chloro) methylidene acetohydrazide (5d): 1859 (C=O amide str), 3186 (N-H iminestr), 2854 (C-H str), 1620 (N=C imineStr, 1226 (C-O str), 1342 (C-N Str), 871(C-Cl str),1HNMR (400 MHz, DMSO-d6) δ 11.01(1H, NH), 8.52 (2H, NCH2), 7.26-8.20(4H, Ar-H, Btz), 6.67-7.56(4H, Ar-H), 5.24(1H, CH2), m/z313.05.
2(1H-benzotriazol-1-yl)-N’-(E)-(4-hydroxy-3-methoxyphenyl) methylidene acetohydrazide (5e): 1836 (C=O amide str), 3464 (O-H str), 3163 (N-H iminestr), 2839 (C-H str), 1681 (N=C imineStr, 1226 (C-O str), 1357 (C-N Str), 1280 (C-O-C str).
2(1H-benzotriazol-1-yl)-N’-(E)-(4-hydroxyphenyl) methylidene acetohydrazide (5f): 1828(C=O amide str), 3039 (N-H iminestr), 2831(C-H str), 1678 (N=C imineStr), 1226(C-O str), 1410(C-N Str), 1HNMR (400 MHz, DMSO-d6) δ 9.82 (1H, NH), 7.24 (2H, NCH2), 6.31-7.04 (4H, Ar-H, Btz), 6.62-5.58(4H, Ar-H), 5.32(1H, CH2), m/z [20].
2(1H-benzotriazol-1-yl)-N’-(1E)-(pentylidene) methylidene acetohydrazide (5g): 1882(C=O amide str), 3117 (N-H iminestr), 2831 (C-H str), 1681 (N=C imineStr), 1350 (C-N Str).
2(1H-benzotriazol-1-yl)-N’-(E)-(4-methylphenyl) methylidene acetohydrazide (5h): 1797(C=O amide str), 3186 (N-H iminestr), 2962 (C-H str), 1681 (N=C imineStr).
2(1H-benzotriazol-1-yl)-N’-(E)-(2-furan-2-yl) methylidene acetohydrazide(5i): 1774 (C=O amide str), 3171 (N-H iminestr), 2901 (C-H str),1689 (N=C imineStr), 1280 (C-O str), 3055 (HC=CH Str), 1HNMR (400 MHz, DMSO-d6) δ 10.57(1H, NH), 8.25 (2H, NCH2), 7.36-8.00 (4H, Ar-H, Btz), 6.63-7.26(3H, Furan-H), 5.56 (1H, CH2), m/z 269.06.
In silico ADMET analysis
A very big challenge in drug development is the drugs ADMET properties in humans. In the drug development process, pharmacokinetic and toxicity risks are evaluated in silico to assist choose the best compounds for screening and minimize the time, expense and acceptance rates. Prior to the designing of experiments in silico screening significantly reduces the number of pharmacokinetics-related failures in the clinical phase. Lipinski and coworkers presented the Rule of five (Ro5) in 1997 which determine the important parameters like lipophilicity and solubility. Molinspiration server supports for calculation of important molecular properties such as LogP, polar surface area, number of hydrogen bond donors, and acceptors. Other criteria are given by Veber et al, is the number of Rotatable bonds (nRotb) and Polar Surface Area (PSA) [21-23].
Swiss ADME and PreADME study was used to filter for compounds with favourable Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) properties.
The rules and violation of drug likeness and physicochemical properties is summarized in Table 2. As per the findings all the synthesized derivatives follow the drug likeness rules with no violation.
| Comp ID | CMC like rule | CMC like rule violations | Rule of five | Ghose violations | Veber violations | Rotatable bonds | H-bond acceptors | H-bond donors | MR | TPSA | iLOGP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5a | Qualified | 0 | Suitable | 0 | 0 | 5 | 5 | 2 | 81.6 | 92.4 | 1.6 |
| 5b | Qualified | 0 | Suitable | 0 | 0 | 6 | 4 | 1 | 89.51 | 72.17 | 2.49 |
| 5c | Qualified | 0 | Suitable | 0 | 0 | 6 | 5 | 1 | 86.07 | 81.4 | 2.56 |
| 5d | Qualified | 0 | Suitable | 0 | 0 | 5 | 4 | 1 | 84.59 | 72.17 | 2.34 |
| 5e | Qualified | 0 | Suitable | 0 | 0 | 6 | 6 | 2 | 88.09 | 101.63 | 2.27 |
| 5f | Qualified | 0 | Suitable | 0 | 0 | 5 | 5 | 2 | 81.6 | 92.4 | 1.77 |
| 5g | Qualified | 0 | Suitable | 0 | 0 | 7 | 4 | 1 | 74.32 | 72.17 | 2.31 |
| 5h | Qualified | 0 | Suitable | 0 | 0 | 5 | 4 | 1 | 84.54 | 72.17 | 2.12 |
| 5i | Qualified | 0 | Suitable | 0 | 0 | 5 | 5 | 1 | 71.84 | 85.31 | 1.77 |
Table 2: Drug likeness and physicochemical properties of benzotriazole N-substituted acetohydrazide derivatives.
The outcome of in sillico ADMET studies shows that the derivatives has significant pharmacological properties like high GI absorption with less or negligible BBB penetration, significant protein binding (more than 90%) and moderate mutagenicity as depicted in Table 3. Screened compounds were within the PSA limits and showed optimum cell permeability and good absorption.
| Comp code | BBB | CaCo2++ Cell permeability | HIA | MDCK | PPB | Algae at | Ames test | Carcino rat | hERG inhibition |
|---|---|---|---|---|---|---|---|---|---|
| 5a | 0.170 | 15.92 | 93.95 | 5.92 | 96.61 | 0.038 | Mutagen | Negative | Low risk |
| 5b | 0.090 | 7.84 | 96.08 | 187.55 | 100 | 0.052 | Mutagen | Negative | Low risk |
| 5c | 0.442 | 5.12 | 96.63 | 77.14 | 100 | 0.037 | Mutagen | Negative | Medium risk |
| 5d | 0.556 | 19.37 | 96.11 | 144.62 | 97.52 | 0.028 | Mutagen | Negative | Medium risk |
| 5e | 0.087 | 15.74 | 94.20 | 15.66 | 94.68 | 0.032 | Mutagen | Negative | Medium risk |
| 5f | 0.255 | 6.84 | 93.96 | 43.65 | 98.45 | 0.042 | Mutagen | Negative | Medium risk |
| 5g | 0.389 | 3.55 | 95.94 | 249.37 | 92.3 | 0.054 | Mutagen | Negative | Medium risk |
| 5h | 0.240 | 11.33 | 96.10 | 192.68 | 100 | 0.033 | Mutagen | Negative | Medium risk |
| 5i | 0.400 | 3.53 | 96.16 | 275.80 | 93.52 | 0.084 | Mutagen | Negative | Medium risk |
Table 3: ADMET data of benzotriazole N-substituted acetohydrazide derivatives.
Molecular docking
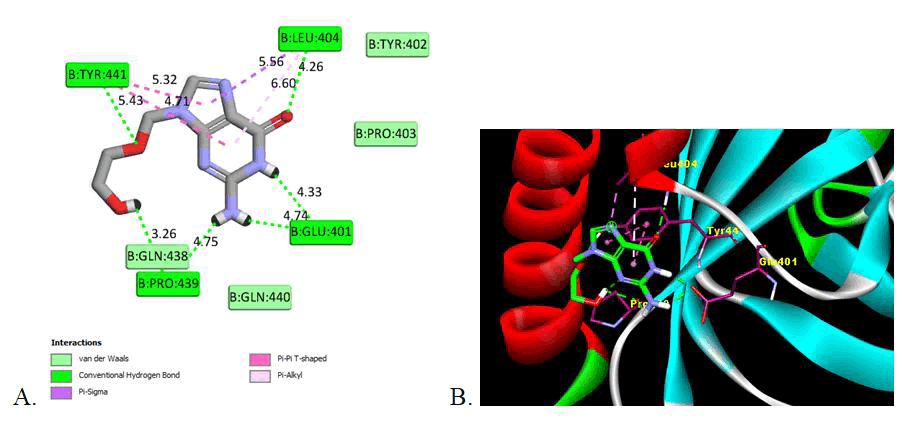
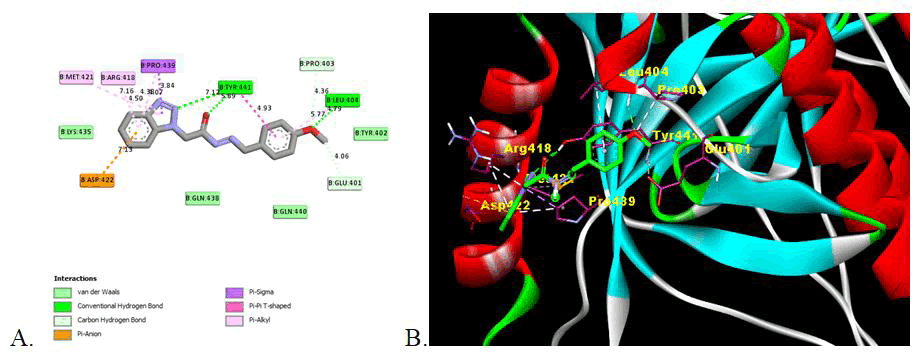
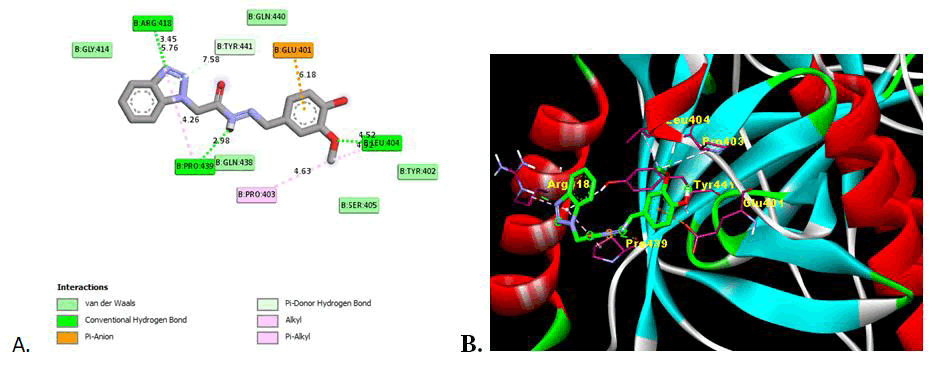
In this study, the inhibitory effect of synthesized derivatives against the glycoprotein B of Herpes Simplex Virus-I RBD crystal structure (PDBID: 2GUM) was investigated using molecular modelling. The docking carried out in AutoDock 4.2 and discovery studio was used to obtain details of interactions like amino acid involved, type of interaction and hydrogen bonding with the amino acid in the active domain (Table 4).
| Comp | Binding energy (kcal/mol) | RMSD | Inhibition constant (Ki, µM) | No. of H-bond (Drug enzyme) | Amino acid involved in interaction | Type of interaction | Amino acids |
|---|---|---|---|---|---|---|---|
| Reference (Acyclovir) | -5.57 | 136.82 | 82.22 | 6 | Glu401 (4.33, 4.74), Leu404 (4.26), Pro439 (3.26, 4.75), Tyr441 (5.43) | Van der Waals, Conventional Hydrogen Bond, Pi-Sigma, Pi-Pi-T-shaped, Pi-Alkyl | Glu401, Tyr402, Pro403, Leu404, Gln438, Pro439, Gln440, Tyr441 |
| 5a | -6.12 | 134.99 | 32.83 | 3 | Pro439 (3.97), Tyr441 (6.71, 5.43) | Van der Waals, Conventional Hydrogen Bond, Pi-Anion, Pi-Alkyl | Glu401, Leu404, Gly414, Ala417, Arg418, Met421, Asp422, Lys435, Gln438, Pro439, Gln440, Tyr441 |
| 5b | -6.27 | 135.58 | 25.19 | 2 | Try441 (5.66,6.80) | Van der Waals, Conventional Hydrogen Bond, Pi-Anion, Pi-Sigma, Pi-Alkyl | Glu401, Tyr402, Pro403, Leu404, Ala417, Arg418, Met421, Asp422, Lys435, Gln438, Pro439, Tyr441 |
| 5c | -6.24 | 135.71 | 26.74 | 3 | Leu404 (4.79), Tyr441 (5.89, 7.12) | Van der Waals, Conventional Hydrogen Bond, Carbon Hydrogen Bond, Pi-anion, Pi-Sigma, Pi-Pi-T-Shaped, Pi-Alkyl | Glu401, Tyr402, Pro403, Leu404, Arg418, Met421, Asp422, Lys435, Gln438, Pro439, Gln440, Tyr441 |
| 5d | -6.21 | 135.49 | 27.99 | 2 | Tyr441 (5.27, 6.62) | Van der Waals, Conventional Hydrogen Bond, Pi-anion, Pi-Alkyl | Glu401, Leu404, Gly414, Ala417, Arg418, Met421, Asp422, Lys435, Gln438, Pro439, Gln440, Tyr441 |
| 5e | -6.44 | 148.14 | 19.09 | 3 | Leu404 (4.52), Arg418 (3.45), Pro439 (2.98) | Van der Waals, Conventional Hydrogen Bond, Pi-Donor Hydrogen Bond, Pi-anion, Alkyl, Pi-Alkyl | Glu401, Tyr402, Pro403, Leu404, Ser405, Gly414, Arg418, Gln438, Pro439, Gln440, Tyr441 |
| 5f | -5.97 | 132.72 | 41.90 | 3 | Glu401 (3.80), Tyr441 (5.36, 6.80) | Van der Waals, Conventional Hydrogen Bond, Pi-anion, Pi-Sigma, Pi-Alkyl | Glu401, Leu404, Gly414, Ala417, Arg418, Met421, Asp422, Lys435, Gln438, Pro439, Gln440, Tyr441 |
| 5g | -5.81 | 132.94 | 55.35 | 2 | Tyr441 (4.88, 6.69) | Van der Waals, Conventional Hydrogen Bond, Carbon, Carbon Hydrogen Bond, Pi-anion, Amide-Pi-Stacked, Pi-Alkyl | Glu401, Gly414, Ala417, Arg418, Met421, Asp422, Lys435, Gln438, Pro439, Gln440, Tyr441 |
| 5h | -6.24 | 132.42 | 26.77 | 1 | Tyr441 (6.56) | Van der Waals, Conventional Hydrogen Bond, Carbon Hydrogen Bond, Pi-Donor Hydrogen Bond, Pi-anion, Pi-Sigma, Pi-Sulfur, Pi-Pi T- shaped, Alkyl, Pi-Alkyl | Glu401, Tyr402, Pro403, Leu404, Gly414, Ala417, Arg418, Met421, Asp422, Lys435, Gln438, Pro439, Gln440, Tyr441, Gln453 |
| 5i | -5.38 | 134.49 | 113.36 | 1 | Pro439 (3.32) | Van der Waals, Conventional Hydrogen Bond, Carbon Hydrogen Bond, Pi-anion, Alkyl, Pi-Alkyl | Glu401, Tyr402, Pro403, Leu404, Gly414, Ala417, Arg418, Met421, Gln438, Pro439, Gln440, Tyr441, Ala451 |
Table 4: Binding energies and interactions of synthesized benzotriazole N-substituted acetohydrazide derivatives against glycoprotein B of herpes simplex virus-I (PDBID: 2GUM).
According to the theoretical affinity values calculated by AutoDock Vina 1.1.2, 204 molecules from three databases were submitted to a detailed visual inspection. During the visual inspection, the binding mode and the interactions with important residues of the active site were considered, such as the hydrogen bond with GLU 401, LEU 404, ARG 418, PRO 439 and TYR441 of gB glycoprotein in active domain. Other interaction with the various amino acids is Van der Waals, conventional hydrogen bond, Pi-Anion, Pi-Alkyl.
All the synthesized derivatives show the significant binding affinities with the amino acid in domain II of gB glycoprotein of HSV-I with binding affinities from -5.38 kcal/mol to -6.44 kcal/mol as compare to -5.57 kcal/mol for reference drug (Acyclovir). Except compound 5i all other has strongest binding affinity that is potent inhibition of glycoprotein. Compound 5e shows the strong inhibition (-6.44 kcal/mol) of gB glycoprotein among the synthesised series. The presence of hydroxy and methoxy group may influence the binding with the amino acid with the strong hydrogen binding hence showing the more inhibition activity. Compound 5c also show higher inhibition with the binding energy -6.24 also has methoxy substituent.
This indicates that the presence of electron withdrawing substituent shows the potent inhibition activity as compare to electron donor groups. The two Dimensional (2D) and three Dimensional (3D) docking images are shown in Figures 2-4 for the reference drug and compound showing the lowest binding energy [24].
Figure 2: Image of molecular docking interaction of reference drug (Acyclovir) with the gB glycoprotein of herpes simplex virus-I (PDBID: 2GUM). A) 2D interaction showing the amino acid involved, type of interaction and the bonding distance, B) 3D interaction of most stable confirmation of ligand showing the bonding with amino acid in active domain.
Figure 3: Image of molecular docking interaction of compound 5c with the gB glycoprotein of herpes simplex virus-I (PDBID: 2GUM). A) 2D interaction showing the amino acid involved, type of interaction and the bonding distance, B) 3D interaction of most stable confirmation of ligand showing the bonding with amino acid in active domain.
Figure 4: Image of molecular docking interaction of compound 5e with the gB glycoprotein of herpes simplex virus-I (PDBID: 2GUM). A) 2D interaction showing the amino acid involved, type of interaction and the bonding distance, B) 3D interaction of most stable confirmation of ligand showing the bonding with amino acid in active domain.
CONCLUSION
Herpes Simplex Virus 1 (HSV-1) is important complex virus involved in the infection. Benzotriazole N-substituted acetohydrazide derivatives show the significant inhibitory activity on glycoprotein B. Compound 5e shows the significant inhibitory activity among the all derivatives. The more stringent structure correlation with the inhibitory activity can be carried out to identify possible modification in synthesised benzotriazole N-substituted acetohydrazide derivatives to modify the inhibition activity significantly. The substitution of electron withdrawing groups can further improve the binding and thereby the inhibitory activity.
References
- Brito MA. Braz J Pharm Sci. 2011;47(4):797-805.
- Chinchole PP, Wankhede SB. Res J Pharm Technol. 2019;12(10):4973-4980.
- Chowdary TK, Cairns TM, Atanasiu D, et al. Nat Struct Mol Biol. 2010;17(7):882-888.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V, et al. Sci Rep. 2016;7:42717.
[Crossref] [Google Scholar] [PubMed]
- Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-250.
[Crossref] [Google Scholar] [PubMed]
- Farooq AV, Shukla D. Surv Ophthalmol. 2012;57(5):448-462.
[Crossref] [Google Scholar] [PubMed]
- Fatahzadeh M, Schwartz RA. J Am Acad Dermatol. 2007;57(5):737-763.
[Crossref] [Google Scholar] [PubMed]
- Gadhave RV, Vichare V, Joshi SV. Asian J Res Chem. 2012;5(7):918-921.
- Ghannam IA, Ali IH, Sheir DH, et al. Bioorg Chem. 2019;93:103373.
[Crossref] [Google Scholar] [PubMed]
- Kawada JI. Adv Exp Med Biol. 2018;1045:85-102.
[Crossref] [Google Scholar] [PubMed]
- Kumar A, De S, Moharana AK. Virol J. 2021;18(1):103.
[Crossref] [Google Scholar] [PubMed]
- Laskowski RA, Swindells MB. J Chem Inf Model. 2011;51(10):2778-2786.
[Crossref] [Google Scholar] [PubMed]
- Li XX, Chen Z. Acta Crystallogr Sect E Struct Rep Online. 2011;67(1):140.
[Crossref] [Google Scholar] [PubMed]
- Lipinski CA. Drug Discov Today Technol. 2004;1(4):337-341.
[Crossref] [Google Scholar] [PubMed]
- Banck M, James CA, Morley C, et al. J Cheminform. 2011;3(33):1-14.
[Crossref] [Google Scholar] [PubMed]
- Patil A, Chaurasia G. J Pharm Sci Res. 2013;5(1):1-4.
- Ranjith D, Ravikumar C. J Pharmacogn Phytochem. 2019;8(5):2063-2073.
- Sangeetha K, Saiz JC, Meena KS, et al. Microb Pathog. 2020;149:104540.
[Crossref] [Google Scholar] [PubMed]
- Trott O, Olson AJ. J Comput Chem. 2010;31(2):455-461.
[Crossref] [Google Scholar] [PubMed]
- Lavinya BU, Sangeetha N, Manisha P, et al. Res J Pharm Technol. 2018;11(2):753-757.
- Veber DF, Johnson SR, Cheng HY, et al. J Med Chem. 2002;45(12):2615-2623.
[Crossref] [Google Scholar] [PubMed]
- Viegas DJ, Edwards TG, Bloom DC, et al. Antiviral Res. 2019;172:104621.
[Crossref] [Google Scholar] [PubMed]
- Wallace AC, Laskowski RA, Thornton JM. Protein Eng. 1995;8(2):127-134.
[Crossref] [Google Scholar] [PubMed]
- Wu J, Power H, Valtchev P, et al. Pharmaceuticals. 2022;15(3):361.
[Crossref] [Google Scholar] [PubMed]