Case Reports: 2020 Vol: 12 Issue: 12
Potential Inhibitor of SARS-CoV-2 main Protease from Andrographis Paniculata: an in Silico Approach
Abstract
Corona virus disease is caused by SARS-COV2 and represents the causative agent of a potentially fatal disease that is of great global public health concern. Corona virus disease has infected about 12,102,328 patients and brought forth death rate about 551,046 among 213 countries and territories as mentioned by WHO in the month of July 2020. There is an emerging need to search for potential medications. The current study aimed to assess bioactive compound found in Andrographis paniculata as potential inhibitors of SARS-COV2, using a molecular docking study. Molecular docking was performed using Autodock 4.2, with the Lamarckian Genetic Algorithm, to analyze the probability of docking. SARS-CoV-2 main protease was docked with bioactive compounds of Andrographis paniculata. Hydroxy chloroquine, chloroquine, remdesivir were used as standards for comparison. The binding energies obtained from the docking of SARS-CoV-2 main protease with andrographolide, neoandrograpolide, andrograpanin, 14-deoxyandrographolide, hydroxy chloroquine, chloroquine, remdesivir were -8.62, -6.93, -7.16, - 8.03, -5.26, -6.37, -3.8 kcal/mol, respectively. These molecules also obey the Lipinski’s rule and computed ADME profile showed that the hits from our study are safe and drug-like compounds. Since it is from a natural source the compound is nontoxic and has reduced side effects.
Keywords
SARS-Cov-2, Molecular Docking, Andrographis Paniculata, ADME
Introduction
Novel corona virus disease (COVID-19) has become a pandemic threat to the public health [1]. The entire world in late December 2019 witnessed a sudden outbreak of COVID-19 that started in Wuhan city of Hubei province of China and then got spread worldwide, rapidly (Chen et al., 2020, Kupferschmidt & Cohen, 2020, Lai et al., 2020) [2]. The epidemic of Covid-19 was declared a pandemic on 12th March 2020 by the world health organization (WHO) (Zhang et al., 2020). It is caused by the virus known as SARS-CoV-2 (previously called 2019-nCoV). This name was chosen because the virus is related to the corona virus that caused the SARS outbreak of 2003; can cause symptoms such as fever, cough, pneumonia, nausea, and fatigue [3-4]. These symptoms are usually mild and begin gradually. Some people become infected, but only have very mild symptoms. Most people (about 80%) recover from the disease without needing hospital treatment. Around 1 out of every 5 people who gets COVID-19 becomes seriously ill and develops difficulty breathing. Older people, and those with underlying medical problems like high blood pressure, heart and lung problems, diabetes, or cancer, are at higher risk of developing serious illness [5]. As of now SARS-CoV-2 has reached 213 countries and territories around the globe, with more than 12,102,328 cases confirmed as of July 10, 2020[6].Around the globe, researchers are endeavoring for the development of effective prevention and treatment strategies for COVID-19. Preliminary studies have suggested potential repurposing of available anti-viral drugs like protease inhibitors lopinavir/ritonavir, nucleoside analogues, neuraminidase inhibitors, remdesivir, umifenovir, tenofovir disoproxil, and lamivudine, etc. for the treatment of COVID-19 infected patients [7]. Recently, Khaerunnisa et al, reported Mpro Inhibitory potential of kaempferol, quercetin, luteolin-7- glucoside, demethoxycurcumin, naringenin, apigenin-7-glucoside, oleuropein, curcumin, catechin, and epicatechin-gallate against COVID-19 [8]. Currently, there are still no effective treatments that target the corona virus and the development of these treatments requires months and years.
Plant compounds are an ideal of finding drug components of interest and most economical one to produce quickly as possible. This is known as the concept of repurposing the natural phytomolecules which will hasten the drug discovery process. Andrographis paniculata is one of the most valuable medicinal plant and bio-factory of diterpronidlactons which have immense value like anticancer [9], anti-diarrheal, anti-hepatitis, anti-HIV, antihyperglycemic, anti-inflammatory, antimicrobial, anti-malarial, antioxidant, cardiovascular, cytotoxic, hepatoprotective, immune-stimulatory, anti- fertility and sexual dysfunctions and bile secretion stimulating agent [10]. A number of active components are reported in this plant which mainly includes diterpone lactones, flavonoides and polyphenols [11].Andrographolide was found to have antiviral properties over many types of viral infections (Gupta et al., 2017) and was found to have activity against Chikungunya (Wintachai et al., 2015). Also, during the outbreak of dengue in India at 2006, aqueous extracts of Andgrographis paniculata were given through the advice of Ministry of AYUSH (Ayurveda, Unani, Siddha and Homeopathy) department of India which led to decrease in cases and infection of the disease as a preventive measure even to normal people acting as an immune booster. The study of anti-dengue activity was found and published through quantification of dengue viral inhibition and showed most antiviral inhibitory effects when DENV 1-4 infected Vero cells with maximum non-toxic dose (Krishnasamy et al., 2018) [12]. Andrographolide, neoandrographolide and 14-deoxy-11,12- didehydroandrographolide were reported to have an anti-HSV-1 activity (Seubsasana S et al., 2011) [13]. Due to these antiviral activities the Andrographis paniculata was chosen, but no reports are available for anti-viral property for corona virus disease.
Hence, the current study aimed to assess bioactive compound found in Andrographis paniculata as potential inhibitors of COVID-19, using a molecular docking study. The molecular docking study makes it easy to identify the inhibitory property of active compounds against many dreadful and chronic diseases. This is because of it reduces the efforts for clinical trial studies or in vivo studies. Here the molecular docking study of bioactive compound of Andrographis paniculata against SARS-COV2 main protease has been carried out and the findings of the present study will provide other researchers with opportunities to identify the right drug to combat COVID-19.
Case Presentation
Target preparation
Crystal structures of target proteins of SARS-CoV-2 main protease (PDB ID: 6Y84) was downloaded from the online protein structure repository, Protein Data Bank (PDB).
Ligand preparation
The Structure of compounds of Andrographis paniculata such as Andrographolide (PubChem CID: 5318517), Neoandrograpolide (PubChem CID: 9848024), Andrograpanin (PubChem CID: 11666871), 14- deoxyandrographolide (PubChem CID: 11624161), and standard drugs such as Hydroxy chloroquine (PubChem CID: 3652), chloroquine (PubChem CID: 2719), Remdesivir (PubChem CID: 121304016) were downloaded from Pubchem in SDF format and converted to PDB format. The compound structures were energy minimized and considered for docking studies.
Molecular Docking
Molecular docking is a method to confirm the binding mode and interaction energy for the ligands with the target protein. Automated docking was performed using Autodock 4.3.
ADME Prediction
ADME (Adsorption, Distribution, Metabolism and Excretion) is important to analyze the pharmacodynamics of the proposed molecule which could be used as a drug. SWISS-ADME is online tool (http://www.swissadme.ch/) which allows the user to draw their respective drug molecule or enter a SMILES of chemical constituents and click run and provides the parameters such as lipophilicity, water solubility, drug likeness and medicinal chemistry (Daina et al., 2017)[14]. The data from PubChem (https://pubchem.ncbi.nlm.nih.gov/) which consists of SMILES of andrographolide, neoandrograpolide, andrograpanin and 14-deoxyandrographolide were entered into the search bar and the parameters were analyzed.
Results and Discussion
Molecular Docking
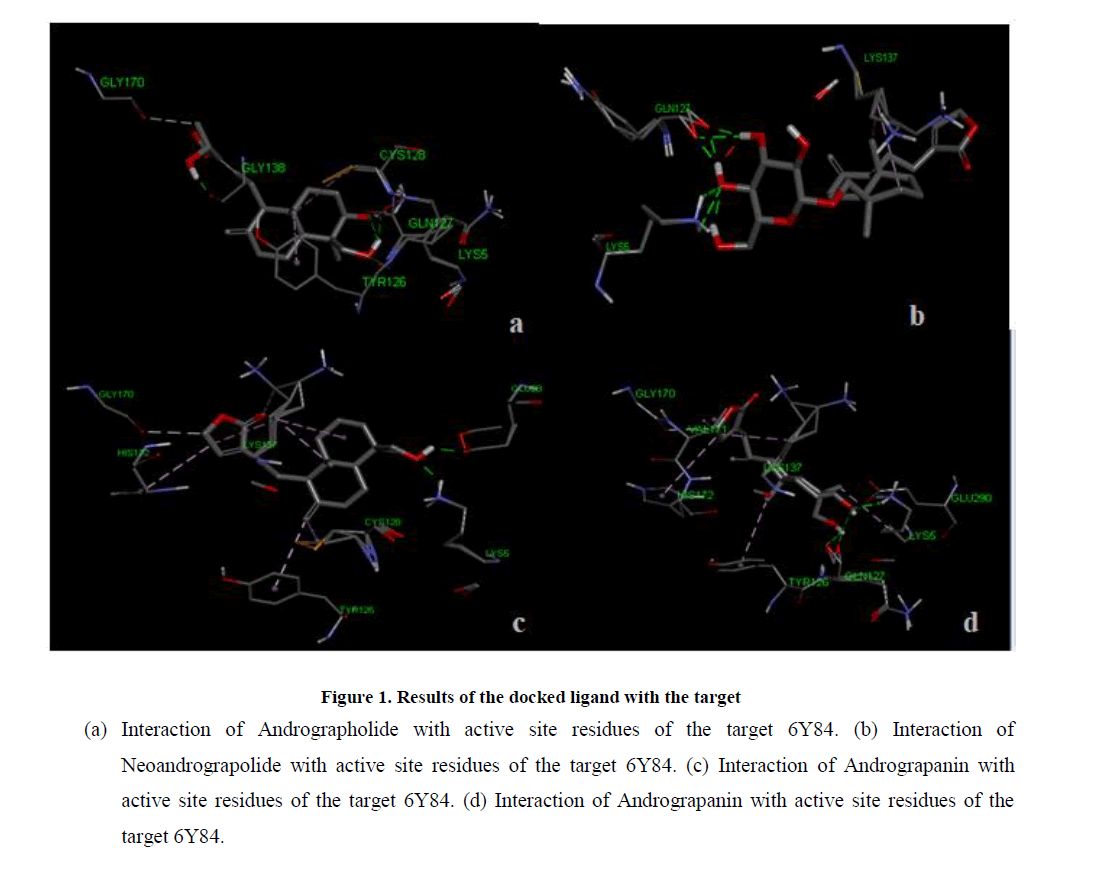
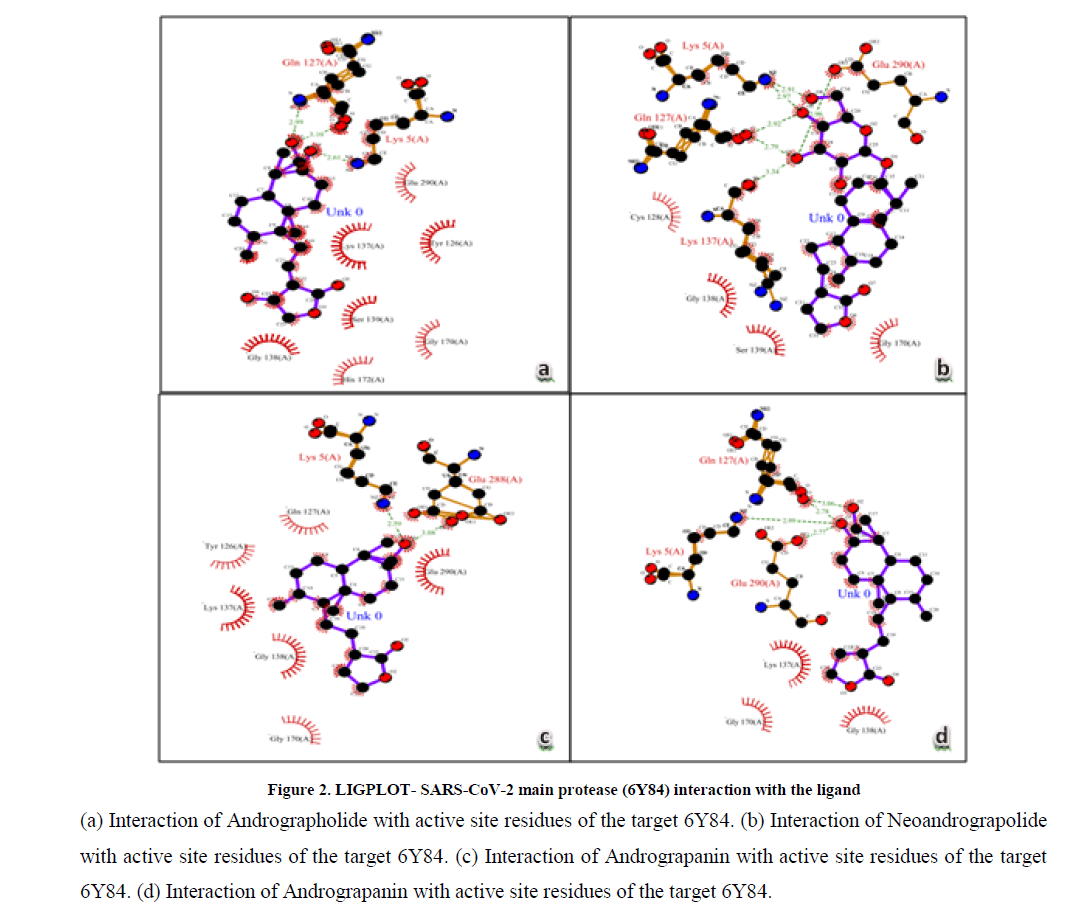
Molecular docking of target protein SARS-CoV-2 main protease was performed with ligands using automated docking protocol. The interaction of protein and ligands for the docked ligands with least binding energy was calculated. Analysis of the receptor/ligand complex models generated after successful docking of the ligands, based on the parameters such as, hydrogen bonds distance, amino acids interactions, binding energy and orientation of the docked bioactive compound with the active site. Andrographolide (-8.62 kcal/mol) and 14-deoxyandrographolide (- 8.03 kcal/mol) showed least binding energy with SARS-CoV-2 main protease. It indicates that bioactive compound of Andrographolide and 14-deoxyandrographolide shows the higher binding energy for target proteins. Moreover in Andrographolide and 14-deoxyandrographolide exhibited good interactions with SARS-CoV-2 main protease. The interaction of SARS-CoV-2 main protease with Andrographis paniculata active compounds shown in Table 1, Figure 1 and Figure 2; and the interaction of SARS-CoV-2 main protease with standard drugs shown in Table 2.
| Target protein | Active Components | Binding energy (kcal/mol) | Interaction with amino acids residues |
|---|---|---|---|
| SARS-CoV-2 main protease (PDB ID: 6Y84) | Andrographolide | -8.62 | GLY170, GLY138, TYR126, LYS5, CYS128, GLN127 |
| Neoandrograpolide | -6.93 | LYS137, GLN127, LYS5 | |
| Andrograpanin | -7.16 | GLY170, HIS172,LYS137, CYS128, TYR126, LYS5, GLU288 | |
| 14-deoxyandrographolide | -8.03 | GLY170, VAL171, HIS172, TYR126, GLN127, LYS5, GLU290, LYS137 |
Table 1. Docking of SARS-CoV-2 main protease with active compounds
| Target protein | Active Components | Binding energy (kcal/mol) | Interaction with amino acids residues |
|---|---|---|---|
| SARS-CoV-2 main protease (PDB ID: 6Y84) | Hydroxy chloroquine | -5.26 | LYS137, GLY138, GLN127, LYS5, TYR126 |
| Chloroquine | -6.37 | VAL171, HIS172, LYS137, TYR126, LYS5, GLU290 | |
| Remdesivir | -3.8 | LYS137, GLU288, GLN127, LYS5 |
Table 2: Docking of SARS-CoV-2 main protease with standard drugs
Figure 1: Results of the docked ligand with the target
(a) Interaction of Andrographolide with active site residues of the target 6Y84. (b) Interaction of Neoandrograpolide with active site residues of the target 6Y84. (c) Interaction of Andrograpanin with active site residues of the target 6Y84. (d) Interaction of Andrograpanin with active site residues of the target 6Y84.
Figure 2: LIGPLOT- SARS-CoV-2 main protease (6Y84) interaction with the ligand
(a) Interaction of Andrographolide with active site residues of the target 6Y84. (b) Interaction of Neoandrograpolide with active site residues of the target 6Y84. (c) Interaction of Andrograpanin with active site residues of the target 6Y84. (d) Interaction of Andrograpanin with active site residues of the target 6Y84.
ADME Prediction
Lipinski's rule of five states that, in general, an orally active drug has not more than 5 hydrogen bond donors (OH and NH groups), not more than 10 hydrogen bond acceptors (notably N and O), molecular weight under 500 g/mol, partition coefficient log P less than 5, number of violation less than 4 [14]. All the compounds were found in compliance with Lipinski's rule of five and the results are reported in Table 3-6.
| Compound | MW (g/mol) | TPSA (Ų) | HBA | HBD | RB | MLogP | Water solubility |
|---|---|---|---|---|---|---|---|
| Andrographolide | 350.45 | 86.99 | 5 | 3 | 3 | 1.98 | Soluble |
| Neoandrographolide | 480.59 | 125.68 | 8 | 4 | 7 | 1.26 | Moderately soluble |
| Andrograpanin | 318.45 | 46.53 | 3 | 1 | 4 | 3.66 | Moderately soluble |
| 14-deoxyandrographolide | 334.45 | 66.76 | 4 | 2 | 4 | 2.81 | Soluble |
MW: Molecular weight; TPSA: Topological polar surface area; HBA: Number of hydrogen bond acceptor; HBD: Number of hydrogen bond donors; RB: Number of ratable bonds; M Log P: Partition coefficient
Table 3: Physicochemical properties
| Compound | GI | BBB Permeant |
P-gp substrate |
Inhibitors | Log Kp (cm/s) | ||||
|---|---|---|---|---|---|---|---|---|---|
| CYP 3A4 |
CYP |
CYP 2C19 |
CYP 2C9 |
CYP D6 |
|||||
| Andrographolide | High | No | Yes | No | No | No | No | No | -6.90 |
| Neoandrographolide | High | No | Yes | Yes | No | No | No | No | -7.36 |
| Andrograpanin | High | Yes | No | No | No | Yes | Yes | No | -5.25 |
| 14-deoxyandrographolide | High | Yes | Yes | No | No | No | No | No | -5.90 |
GI: Gastro- Intestinal absorption; BBB permeant: Blood-Brain-Barrier permeant; P-gpsubstrate: Permeability glycoprotein substrate; Log Kp: Skin permeation
Table 4: Pharmacokinetics prediction
| Compound | Lipinski rule | Ghose filter | Veber filter | Egan filter | Muegge filter | Bioavailability Score |
|---|---|---|---|---|---|---|
| Andrographolide | Yes; 0 violation | Yes | Yes | Yes | Yes | 0.55 |
| Neoandrographolide | Yes; 0 violation | No: 2 violations | Yes | Yes | Yes | 0.55 |
| Andrograpanin | Yes; 0 violation | Yes | Yes | Yes | Yes | 0.55 |
| 14-deoxyandrographolide | Yes; 0 violation | Yes | Yes | Yes | Yes | 0.55 |
Table 5: Drug likeness prediction
| Compound | Lead- likeness | PAINS structural alert | Brenk structural alert | Synthetic accessibility score |
|---|---|---|---|---|
| Andrographolide | No; 1 violation | 0 | 2 | 5.06 |
| Neoandrographolide | No; 1 violation | 0 | 1 | 6.36 |
| Andrograpanin | No; 1 violation: | 0 | 1 | 4.63 |
| 14-deoxyandrographolide | Yes | 0 | 1 | 4.82 |
Table 6: Medicinal chemistry prediction
Conclusion
Nowaday’smoleculardockingplayakeyroleinunderstandingdrugreceptorinteraction, which further help in designing novel, or potent inhibitors through drug receptor interaction mechanism. The pandemic of COVID–19 is an unprecedented global public health challenge to develop effective drugs for prevention and treatment with minimum or no side effects. Traditional Indian medicinal plants have long been used for treatment of several diseases, including antiviral therapeutics against several viruses. This study observed that bioactive components in Andrographis paniculata displayed better binding affinities than the synthetic drugs currently in use for treatment for COVID–19 infection. Drug likeness factor rules were obeyed accordingly without any violation, which describes the compounds, can act as a drug in the biological systems. Further in-vitro and in-vivo analyses are required to transform these potential inhibitors into clinical drugs.
References
- R Hussin; NB Siddappa. J Autoimmun. 2020, 102433.
- AA Elfiky. Life Sci. 2020, 253, 117-592.
- YR Guo; QD Cao; ZS Hong; YY Tan; SD Chen, HJ Jin. Mil Med Res. 2020, 7(1), 11.
- WHO website: Coronavirus disease(COVID-19)
- Southern Cross Care website: What is COVID-19?
- World Health Organization (WHO), Coronavirus disease 2019 (COVID-19). 2020,1-9.
- M Hoffmann; H Kleine-Weber Schroeder; S Kruger. Cell. 2020, 181, 1-10.
- S Khaerunnisa; H Kurniawan; R Awaluddin; S Suhartati; S Soetjipto. Prepr. 2020, 20944, 1-4.
- R Sharmila. IJPSDR. 2013, 5(2), 56-61.
- S Hossain; Z Urbi; A Sule; KM Rahman. Photochemistry and pharmacology Sci World J. 2014.
- S Akbar. Alter Med Rev. 2011, 16(1), 66–77.
- A Enmozhi; SK Raja; K Sebastine; J Joseph. J Biomol Struct Dyn. 2020, 1-10.
- S Seubsasana; C Pientong; T Ekalaksananan; S Thongchai; C Aromdee. Med Chem. 2011, 7(3), 237-244.
- A Daina; O Michielin; V Zoete. Sci Rep. 2017, 7, 42717.