Original Articles: 2023 Vol: 15 Issue: 8
A Systemic Review on Bioinformatics Tools and Approaches to Combat the Antibiotic Resistance
Mulagani P Evangelin1*, Prem Kumar P2, Balamurugan K1
1Department of Pharmacy, Annamalai University, Tamil Nadu, India
2Department of Pharmacy, Mangaldas University, Guntur, India
- Corresponding Author:
- Mulagani P Evangelin
Department of Pharmacy,
Annamalai University,
Tamil Nadu,
India.
Received: 20-Apr-2020, Manuscript No. JOCPR-23-9474; Editor assigned: 23-Apr-2020, PreQC No. JOCPR-23- 9474 (PQ); Reviewed: 07-May-2020, QC No. JOCPR-23-9474; Revised: 19-Jul-2023, Manuscript No. JOCPR-23- 9474 (R); Published: 16-Aug-2023
Citation: Evangelin MP, et al. 2023. Novice Cinnoline Derivatives as Antibacterial Agent and Antimycobacterial Agent: Synthesis, Microbial Evaluation and Molecular Docking Study. J. Chem Pharm. Res., 15:042.
Copyright: © 2023 Evangelin MP, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Antibiotic resistance is a significant issue worldwide as overexposure of antibiotics in the form of excessive and unauthorized prescription by physicians or other medical personnel, the antibiotics added in food and fodder of animals in agriculture and aquaculture, etc. took a crucial role in the development of resistance genes against antibiotics as we can easily withstand by microbial residents of the soil, water. Our microbiota possesses such genes that can resist the activity or destroy the administered antibiotic molecules entirely and gradually turns into a serious matter. Hence, it is very essential to cultivate new classes of drugs. However, billions of dollars and longer than a decade have been invested in fulfilling this need. But, only a few antibiotics were approved and reported in the past decades. Thus, with the aid of applied bioscience disciplines, humanity has developed a new approach known as bioinformatics, which provided crucial routes for sculpting a natural living cell system and proteins, allowing scientists to discover operational drug strategies to combat the spread of antibiotic resistance among common infectious diseases by examining and elucidating a large amount of data (such as nucleic acid and amino acid patterns, protein domains, protein configurations and protein-ligand solid interaction behavior) at the molecular scale without the use of laboratory studies. Thus, in this article, we will withstand the application of bioinformatics tools and approaches to combat antibiotic resistance with the help of case studies.
Keywords
Cinnoline, Anti-bacterial, Anti tubercular activity, Docking
Introduction
The main challenge in the world now a day is about diseases that are infectious and the pathogens which are resistant to drugs which are known. There are an infinite number of various types of bacteria in the world around where most of them are not harmful to mankind, some of the bacteria cause dangerous diseases of all kinds and a few are panic which leads in chance of death among the elderly so there is a need to synthesize potent antibacterial compounds so our investigation continued in developing a new potent promising antibacterial drug [1]. Antibacterial agents are discovered through this development which inhibits DNA gyrase cinnoline are synthesized and evaluated as antimicrobial agents. Mycobacterium Tuberculosis (MTB) which an intracellular bacterium causes tuberculosis. TB is considered as a global health crisis by the WHO. The main reason for death in the case of TB is due to lack of better treatment to fight against strains of TB. Tuberculosis occupied the third position of death in women and mostly in between the ages of 15-44. Most of the TB cases are from developing states. TB is a dreadful disease in which one-third of the world population is affected by MT [2]. Treatment of TB included multi-drug regimen (isoniazid, rifampicin, pyrazinamide, ethambutol). It is a long and timetaking method which requires continuous monitoring for at least six months. Depending on the upton body immune system the reappearance of symptoms of TB varies from patient to patient. There is a deadly need to synthesize effective drugs with less cost and rapid cure within less period of time [3].
MATERIALS AND METHODS
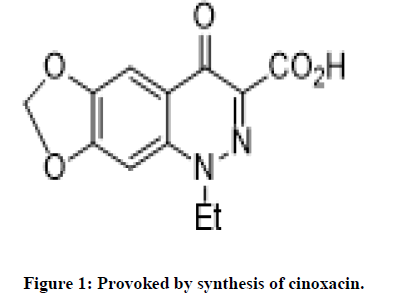
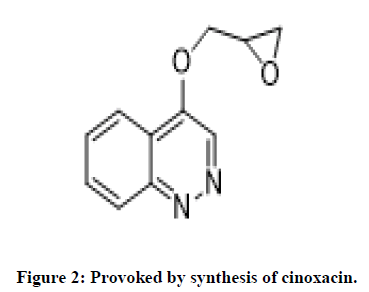
The cinnoline core has antibacterial, antitumor, antifungal, anti-inflammatory activity. Cinnolines also exhibits antituberculosis activity, anesthetizing, sedative activity. Provoked by synthesis of cinoxacin by Helen Giamarellou, evaluation of its antibacterial activity driven our research proposal to synthesize new compounds of scheme-1 with high yield and better potency (Figures 1 and 2) [4].
In drug discovery cinnolines are most commonly used by a slight modification of an already existing one. As per published articles, lipophilicity is the major reason for activity in cinnoline. Many kinds of literature support the activity of cinnoline. In the light of previously published articles, cinnolines are revealed as efficient analog which intrigued us to design and synthesize new derivatives [5]. In addition, cinnolines have been remarkably active against E.coli. In view of the interest in exploited antibacterial activity, cinnolines paved its way towards the research path. These findings envisaged us to construct a novel molecular framework that contains cinnoline ring systems in matrix with the hope of developing a compound that possesses better antibacterial activity.
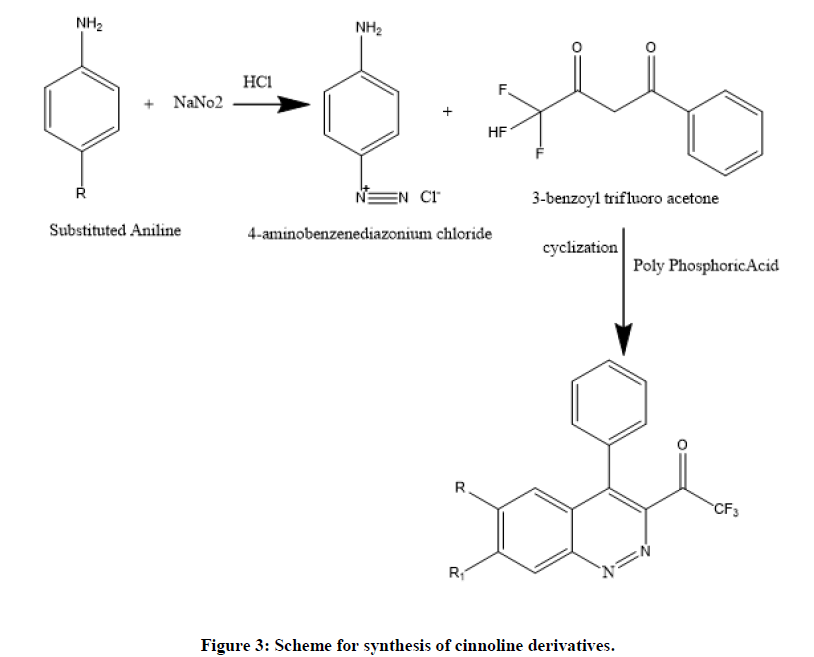
Breakthrough development of cinnoline moiety has intrigued us to synthesis A new series of cinnoline framework and evaluated for antimycobacterial activity and the primary target of isoniazid (INH) is Mycobacterium tuberculosis enoyl-acyl-ACP reductase (InhA). Provoked by synthesis of cinoxacin by Helen Giamarellou at and evaluation of its antibacterial activity driven our research proposal to synthesize new compounds of scheme-1 with high yield and better potency (Figure 3 and Table 1) [6].
| Compound | R | R1 |
|---|---|---|
| CN-1 | NO2 | - |
| CN-2 | NH2 | - |
| CN-3 | CH3 | - |
| CN-4 | Cl | - |
| CN-5 | Br | - |
| CN-6 | I | - |
| CN-7 | COOH | - |
| CN-8 | OH | - |
| CN-9 | SO3H | - |
| CN-10 | SO2NH2 | - |
| CN-11 | Cl | NO2 |
| CN-12 | Cl | CF3 |
| CN-13 | F | Cl |
| CN-14 | OCH3 | - |
Table 1: List of cinnoline compounds.
Chemistry
In these studies, novel cinnoline compounds are designed and synthesized [7]. The strategy of synthesized compounds was given in scheme-1. Afforded compounds CN (1-14) were reacted with different substituted aniline in the presence of sodium nitrite and Hcl. The diazonium salt formed is allowed for cyclization reaction through 3-benzoyl trifluoro acetone in the presence of polyphosphoric acid. The purity of compound checked through TLC and structures of synthesized compounds were confirmed by FTIR and proton NMR (Table 2) [8].
| S.no | Name of the compound (IUPAC) | Molecular formula | Molecular weight | Melting point | % Yield |
|---|---|---|---|---|---|
| 1 | 2,2,2-trifluoro-1-(6-nitro-4-phenylcinnolin-3-yl)ethanone | C16H8F3N3O3 | 347.25 | 159 | 61 |
| 2 | 1-(6-amino-4-phenyl quinolin-3-yl)-2,2,2-trifluoroethanone | C16H10F3N3O | 317.27 | 185 | 66 |
| 3 | 2,2,2-trifluoro-1-(6-methyl-4-phenylcinnolin-3-yl)ethanone | C17H11F3N2O | 316.28 | 197 | 65 |
| 4 | 1-(6-chloro-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone | C16H8ClF3N2O | 336.7 | 212 | 68 |
| 5 | 1-(6-bromo-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone | C16H8BrF3N2O | 381.15 | 221 | 64 |
| 6 | 2,2,2-trifluoro-1-(6-iodo-4-phenylcinnolin-3-yl)ethanone | C16H8F3IN2O | 428.15 | 165 | 57 |
| 7 | 4-phenyl-3-(2,2,2-trifluoroacetyl)cinnoline-6-carboxylic acid | C17H9F3N2O3 | 346.26 | 197 | 65 |
| 8 | 2,2,2-trifluoro-1-(6-hydroxy-4-phenylcinnolin-3-yl)ethanone | C16H9F3N2O2 | 318.25 | 150 | 57 |
| 9 | 4-phenyl-3-(2,2,2-trifluoroacetyl)cinnoline-6-sulfonic acid | C16H9F3N2O4S | 382.02 | 206 | 66 |
| 10 | 4-phenyl-3-(2,2,2-trifluoroacetyl)cinnoline-6-sulfonamide | C16H10F3N3O3S | 381.33 | 221 | 61 |
| 11 | 1-(6-chloro-7-nitro-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone | C16H7ClF3N3O3 | 381.69 | 239 | 67 |
| 12 | 1-(6-chloro-4-phenyl-7-(trifluoromethyl)cinnolin-3-yl)-2,2,2-trifluoroethanone | C17H7ClF6N2O | 404.69 | 169 | 53 |
| 13 | 1-(7-chloro-6-fluoro-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone | C16H7ClF4N2O | 354.69 | 187 | 52 |
| 14 | 1-(7-chloro-6-methoxy-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone | C17H10ClF3N2O2 | 366.72 | 182 | 51 |
Table 2: IUPAC naming of synthesized compounds.
Experimental protocols
Various materials used for synthesis were purchased from respective vendors like sodium nitrate (Merck, Hyderabad, India), 3-benzoyl trifluoro acetone (Merck, Hyderabad, India), (para nitro aniline (Loba Chemie, Mumbai, India), polyphosphoric acid (Otto Chem, Mumbai, India), sulfuric acid (Loba Chemie, Mumbai, India), agar, beeswax, tragacanth gum (Loba Chemie, Mumbai, India). All reagents were analytical grades along with chemicals [9].
Synthesis of 2,2,2-trifluoro-1-(6-substituted-4-phenyl cinnolin-3-yl)ethanone(CN1-14)
Substituted anilines (R1, R2) (0.1 mol) was added to 5 ml of Hcl (200 ml) in cooled condition. To this, sodium nitrite solution was added by stirring while the temperature is maintained below 5°C. To the diazonium salt (0.1 mol) of 3-benzoyl trifluoro acetone and of 2 g of phosphoric acid was allowed to condense for 1 hr [10]. The reaction progress was continuously monitored by TLC and then allowed recrystallization using ethanol and finally, the reaction mixture was added to ice cold water then filtered and dried (scheme-1). Compounds CN (1-14) were prepared by a similar procedure by substituting the R alkyl group. The structures of the compounds (1-14) have been confirmed on the basis of analytical and spectral IR, 1H NMR and mass data. Synthesized compounds properties. Physical properties and IUPAC names are illustrated in Tables 1 and 2 [11].
2,2,2-trifluoro-1-(6-nitro-4-phenylcinnolin-3-yl)ethanone(CN-1): Yield 61%; Mp; 159; IR (KBr, cm-1)1535 (N=N), 800 (C-S), 1609.31 (C=N Stretching), 1385.6 (NO2 stretching), 1601 (C=O), 2862 (CH3), 1215 (C-F), 1021.12 (N-N stretching), 1H NMR (CDCl3) 8.70 (s, 1H, Ar), 8.26 (d, 1H, Ar), 8.53 (d, 1H, Ar), 7.40-7.52 (m, 5H, Ar) m/z: 347.05 C, 55.34; H, 2.32; F, 16.41; N, 12.10; O, 13.82.
1-(6-amino-4-phenyl cinnolin-3-yl)-2,2,2-trifluoroethanone(CN-2): Yield 66%; Mp; 185 IR (KBr, cm1) 3199.33 (NH stretching), 1535(N=N), 800 (C-S), 1609.31(C=N stretching), 1601(C=O), 2862(CH3), 1215(C-F), 1021.12(N-N stretching), 1H-NMR(CDCl3), 7. 92 (d, 1H, Ar), 7.16 (t, 1H, Ar), 6.92 (s, 1H, Ar), 6.25 (s,1H, NH2), 7.40-7.53 (m, 5H, Ar) m/z: 317.08 C, 60.57; H, 3.18; F, 17.96; N, 13.24; O, 5.04.
2,2,2-trifluoro-1-(6-methyl-4-phenylcinnolin-3-yl)ethanone(CN-3): Yield 65%; Mp; 197; IR (KBr, cm; 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601(C=O), 2862(CH3), 1215 (C-F), 1021.12 (N-N Stretching), 1H-NMR(CDCl3)8.01 (d, 1H, Ar), 7.40-7.58 (m, 7H, Ar), 2.32(s, 1H, CH3) m/z: 316.28, C , 64.56; H, 3.51; F, 18.02; N, 8.86; O, 5.06.
1-(6-chloro-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone(CN-4): Yield 68%; Mp; 212; IR(KBr, cm1) 1316.28 (NH stretching), 746 (C-Cl), 1535(N=N), 800 (C-S), 1609.31 (C=N stretching), 1601 (C=O), 2862(CH3), 1215 (C-F), 1021.12 (N-N Stretching) 1H-NMR(CDCl3) 8.01 (d, 1H, Ar), 7.74 (t, 2H, Ar), 7.40-7.53 (m, 5H, Ar), m/z: 336.70, C, 57.08; H, 2.39; Cl, 10.53; F, 16.93; N, 8.32; O, 4.75.
1-(6-bromo-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone(CN-5):Yield 64%; Mp; 221; IR (KBr, cm1) 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601 (C=O), 2862 (CH3), 650 (C-Br), 1215 (C-F), 1021.12 (N-N stretching), 1H-NMR (CDCl3) 4.207.95-8.01(m, 2H, Ar), 7.86 (t, 1H, Ar), 7.40-7.52 (m, 5H, Ar) m/z: 381.25, C, 50.42; H, 2.12; Br, 20.96; F, 14.95; N, 7.35.
2,2,2-trifluoro-1-(6-iodo-4-phenylcinnolin-3-yl)ethanone(CN-6): Yield 57%; Mp; 165; IR (KBr, cm1) 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching),1601 (C=O), 2862 (CH3), 1100 (C-I), 1215 (C-F), 1021.12 (N-N stretching) 1H-NMR (CDCl3) 8.10 (t, 2H, Ar), 7.85 (d, 1H, Ar), 7.40-7.52 (m, 5H, Ar) C16H8F3IN2O, m/z: 428.15, C, 44.88; H, 1.88; F, 13.31; I, 29.64; N, 6.54; O, 3.74.
4-phenyl-3-(2,2,2-trifluoroacetyl)cinnoline-6-carboxylic acid (CN-7): Yield 65%; Mp; 197; IR (KBr, cm1) 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601 (C=O), 2862 (CH3), 1215 (C-F), 1300 (C00H), 1021.12 (N-N stretching) 10.5 (s, 1H, OH), 1H-NMR (CDCl3) 8.61 (s, 2H, Ar), 8.31 (d, 1H, Ar), 7.40-7.52 (m, 5H, Ar) C17H9F3N2O3 m/z: 346.26,C, 58.97; H, 2.62; F, 16.46; N, 8.09; O, 13.86.
2,2,2-trifluoro-1-(6-hydroxy-4-phenylcinnolin-3-yl)ethanone(CN-8): Yield 57%; Mp; 150; 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601 (C=O), 2862 (CH3), 1215 (C-F), 3200 (OH), 1021.12 (N-N stretching) 1H-NMR (CDCl3), 8.05 (d, 1H, Ar), 7.40-7.52 (m, 5H, Ar, 7.03 (s, 1H, Ar), 5.31(s, 1H, OH) C16H9F3N2O2 m/z: 318.25,C, 60.38; H, 2.85; F, 17.91; N, 8.80; O, 10.05.
4-phenyl-3-(2,2,2-trifluoroacetyl)cinnoline-6-sulfonic acid (CN-9): Yield 66%; Mp; 206; IR (KBr, cm-1) 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N Stretching), 1601 (C=O), 2862 (CH3), 1215 (C-F), 1350 (HSO3), 1021.12 (N-N Stretching), 8.36-8.41 (m, 3H, Ar), 7.40-7.52 (m, 5H, Ar), 2.1 (s, 1H, OH), C16H9F3N2O4S, m/z: 382.02 C, 50.27; H, 2.37; F, 14.91; N, 7.33; O, 16.74; S, 8.39.
4-phenyl-3-(2,2,2-trifluoroacetyl)cinnoline-6-sulfonamide(C N-10): Yield 61%; Mp; 221; IR (KBr, cm-1) 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601 (C=O), 2862 (CH3), 1370 (SO2NH2), 1215 (C-F), 1021.12 (N-N Stretching)) 8.41-8.36 (m, 3H, Ar), 2.1 (s, 1H, NH2), 7.40-7.52 (m, 5H, Ar) C16H10F3N3O3S m/z: 381.33 C, 50.40; H, 2.64; F, 14.95; N, 11.02; O, 12.59; S, 8.41.
1-(6-chloro-7-nitro-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone(CN-11): Yield 67%; Mp; 239, 316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N Stretching), 1601 (C=O), 2862 (CH3), 1215 (C-F), 1021.12 (N-N stretching) H-NMR (CDCl3)1(s, 1H, Ar), 7.91 (s, 1H, Ar), 7.40-7.52 (m, 5H, Ar) C16H7ClF3N3O3 m/z: 381.69, C, 50.35; H, 1.85; Cl, 9.29; F, 14.93; N, 11.01; O, 12.58.
1-(6-chloro-4-phenyl-7-(trifluoromethyl)cinnolin-3-yl)-2,2,2-trifluoroethanone(CN-12): Yield 53%; Mp; 169; 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601(C=O), 2862 (CH3), 1215 (C-F), 1021.12 (N-N stretching) 1H-NMR (CDCl3) 8.75 (s, 1H, Ar), 7.96 (s, 1H, Ar), 7.40-7.52 (m, 5H, Ar) C17H7ClF6N2O, m/z:404.69 C, 50.45; H, 1.74; Cl, 8.76; F, 28.17; N, 6.92; O, 3.95.
1-(7-chloro-6-fluoro-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone(CN-13): Yield 52%; Mp; 187; IR (KBr, cm-1) 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601 (C=O), 2862 (CH3), 1215 (C-F), 1021.12 (N-N stretching), 8.35 (d, 1H, Ar), 7.37-7.52 (m, 6H, Ar); C16H7ClF4N2O, m/z: 354.69C, 54.18; H, 1.99; Cl, 10.00; F, 21.43; N, 7.90; O, 4.51.
1-(7-chloro-6-methoxy-4-phenylcinnolin-3-yl)-2,2,2-trifluoroethanone (CN-14): Yield 51%; Mp; 182; IR (KBr, cm-1) 1316.28 (NH stretching), 746 (C-Cl), 1535 (N=N), 800 (C-S), 1609.31 (C=N stretching), 1601 (C=O), 2862 (CH3), 1215 (C-F), 1021.12 (N-N stretching) C17H10ClF3N2O2 m/z: 366.72 C, 55.68; H, 2.75; Cl, 9.67; F, 15.54; N, 7.64; O, 8.731H-NMR (CDCl3) 8.35 (s, 1H, Ar), 7.40-7.52 (m, 5H, Ar), 6.87 (s, 1H, Ar), 3.62 (s, 3H,CH3).
RESULTS AND DISCUSSION
Molecular docking studies
Structure based drug design and molecular studies: Ligand docking studies were performed by Molegro virtual docker (Molegro mApS, Aarhus C and Denmark). Fourteen compounds selected from the search of a new ligand for GyrB ATPase (a domain of DNA Gyrase) inhibitor as a novel antibacterial drug like candidates. The target proteins for docking studies are DNA Gyrase Subunit B (PDB ID: 4BAE). In antimycobacterial activity, the target proteins selected for docking are DHFrase A (PDB ID: 2CIG). The structures were drawn using ChemDraw version 12.0 and saved in mol format after minimization of energy [12]. The 3D structures of target proteins were downloaded from the protein data bank pdb format. Selected chain in the target protein imported into the workspace [13]. Surface was created and binding pockets were predicted and then ligands also imported to the workspace and prepared them for docking. The grid was generated around the binding pocket of co-crystallized ligand and docking was run after setting of parameters such as choosing which ligand to dock, choosing scoring function and defining binding site, choosing search algorithm and number of runs, maximum interaction, maximum population size, energy threshold, maximum steps, neighbour distance factor and pose clustering. The docking score (moldock) of the ligands are compared with co-crystallized ligand of the respective proteins in ciprofloxacin, fin docking results recorded in Tables 3 and 4 [14].
| PDB code | Compounds | Binding energy (Kcal/mol) | Residue involving H-bond |
|---|---|---|---|
| 4BAE | CN-1 | -100.372 | |
| 4BAE | CN-2 | -101.048 | - |
| 4BAE | CN-3 | -93.015 | - |
| 4BAE | CN-4 | -93.844 | - |
| 4BAE | CN-5 | -93.672 | - |
| 4BAE | CN-6 | -118.364 | - |
| 4BAE | CN-7 | -135.44 | Asn 52 |
| 4BAE | CN-8 | -105.21 | - |
| 4BAE | CN-9 | -107.12 | - |
| 4BAE | CN-10 | -101.22 | - |
| 4BAE | CN-11 | -128.534 | Asn 52 |
| 4BAE | CN-12 | -127.469 | Ile 84, hr 169 |
| 4BAE | CN-13 | -101.924 | - |
| 4BAE | CN-14 | -101.821 | - |
Table 3: Molecular docking reports for compounds CN (1-14) against protein DNA Gyrase B.
| PDB code | Compounds | Binding energy (kcal/mol) | Residue involving H-bond |
|---|---|---|---|
| 2CIGDHRase | CN-1 | -138.979 | Ala, Arg, Ser,Val |
| 2CIGDHRase | CN-2 | -128.048 | - |
| 2CIGDHRase | CN-3 | -123.015 | - |
| 2CIGDHRase | CN-4 | -113.844 | - |
| 2CIGDHRase | CN-5 | -121.672 | - |
| 2CIGDHRase | CN-6 | -108.364 | - |
| 2CIGDHRase | CN-7 | -139.382 | Arg, Gly, Gln |
| 2CIGDHRase | CN-8 | -105.21 | - |
| 2CIGDHRase | CN-9 | -107.12 | - |
| 2CIGDHRase | CN-10 | -141.678 | Ala, IIe, Asp, Tyr, Ser - |
| 2CIGDHRase | CN-11 | -139.666 | Gln, Val, Arg |
| 2CIGDHRase | CN-12 | -127.469 | - |
| 2CIGDHRase | CN-13 | -101.924 | - |
| 2CIGDHRase | CN-14 | -101.821 | - |
Table 4: Molecular docking reports for compounds CN (1-14) against protein DHFrase A.
In silico pharmacokinetics (ADME) properties
The designed compounds are predicted for their physicochemical properties using Swiss ADME online software. In general human body, receptor’s pharmacokinetics properties are based on molecular properties. Lipinski introduced this rule for predicting bioavailability of drug like molecule and some physicochemical properties. They are Clog P (1.92 to +5.31), molecular weight (316.28-404.69D), H-bond donors (not more than 1), HBA (not more than 9), rotatable bonds (4 or fewer) polar surface area (equal to or <111.39Å) [19]. Drugs can easily cross the BBB in the log p value between 1.65 and 2.86. According to John the drugs possessing log p value 1.5 to 2.5 can cross the BBB easily [15].
As per in the silico ADME report, all the cinnoline compounds obey Lipinski’s rule of five so these compounds will absorb orally and it can reach its targeted site by crossing BBB (Table 5). It is enforced only to absorption by passive diffusion of title compounds through cell membranes but not to the drugs which absorb through the active transport process.
| Compound code | Molecular weight | Num. rotatable bonds | Num. H-bond acceptors | Num. H-bond donors | TPSA | Log Po/w (iLOGP) | BBB permanent |
|---|---|---|---|---|---|---|---|
| CN-1 | 347.25 g/mol | 4 | 8 | 0 | 88.67 Ų | 2 | No |
| CN-2 | 317.27 g/mol | 3 | 6 | 1 | 68.87 Ų | 2.01 | Yes |
| CN-3 | 316.28 g/mol | 3 | 6 | 0 | 42.85 Ų | 2.69 | Yes |
| CN-4 | 336.70 g/mol | 3 | 6 | 0 | 42.85 Ų | 2.78 | No |
| CN-5 | 381.15 g/mol | 3 | 6 | 0 | 42.85 Ų | 2.85 | No |
| CN-6 | 428.15 g/mol | 3 | 6 | 0 | 42.85 Ų | 2.84 | Yes |
| CN-7 | 346.26 g/mol | 4 | 8 | 1 | 80.15 Ų | 1.95 | No |
| CN-8 | 318.25 g/mol | 3 | 7 | 1 | 63.08 Ų | 2.09 | Yes |
| CN-9 | 382.02 g/mol | 4 | 9 | 1 | 105.60 Ų | 1.65 | No |
| CN-10 | 381.33 g/mol | 4 | 9 | 1 | 111.39 Ų | 1.69 | No |
| CN-11 | 381.69 g/mol | 4 | 8 | 0 | 88.67 Ų | 2.12 | No |
| CN-12 | 404.69 g/mol | 4 | 9 | 0 | 42.85 Ų | 2.87 | No |
| CN-13 | 354.69 g/mol | 3 | 7 | 0 | 42.85 Ų | 2.66 | No |
| CN-14 | 366.72 g/mol | 4 | 7 | 0 | 52.08 Ų | 2.86 | No |
Table 5: In silico ADME properties of cinnoline compounds.
Antimicrobial activity
All the synthesized compounds were evaluated by disk plate method according to standard procedure. Antibacterial activity is screened against Bacillus subtilis MTCC 441, S. aureus ATCC 96, E.coli ATCC 8739, K. pneumoniae MTCC 109 (Table 6). Minimum Inhibitory Concentration (MIC) was determined and tabulated in Table 7. Standard drug used was ciprofloxacin. Experimental results revealed that all the cinnoline candidates have shown activity between range 12.5 μg/ml-100 μg/ml. According to antimicrobial activity result, the reason for the activity in compound-7 is due to presence of carboxylic group which increases the lipophilic nature. In compound-11 electron negative group chlorine enhanced the anti-microbial activity with MIC of 12.5 μg/ml.
| Compounds | B. subtilis | S. aureus | E. coli | K. pneumoniae |
|---|---|---|---|---|
| CN-1 | 12 | 9 | 13 | 12 |
| CN-2 | 10 | 11 | 13 | 14 |
| CN-3 | 12 | 13 | 12 | 13 |
| CN-4 | 10 | 11 | 14 | 13 |
| CN-5 | 12 | 10 | 11 | 12 |
| CN-7 | 13 | 14 | 18 | 17 |
| CN-8 | 12 | 10 | 12 | 13 |
| CN-9 | 12 | 11 | 14 | 11 |
| CN-10 | 10 | 11 | 12 | 13 |
| CN-11 | 13 | 12 | 17 | 16 |
| CN-12 | 12 | 13 | 16 | 15 |
| CN-13 | 11 | 12 | 14 | 11 |
| CN-14 | 12 | 13 | 12 | 13 |
| Ciprofloxacin | 25 | 25 | 25 | 25 |
Gram positive: Bacillus subtilis, Staphylococcus aureus,
Gram negative: Escherichia coli, Klebsiella pneumoniae
Table 6: Antibacterial activity zone of inhibition data (mm).
| CN | B. subtilis | S. aureus | E. coli | K. pneumoniae |
|---|---|---|---|---|
| CN-1 | 100 | 50 | 25 | 50 |
| CN-2 | 50 | 100 | 50 | 100 |
| CN-3 | 100 | 50 | 50 | 100 |
| CN-4 | 50 | 100 | 25 | 50 |
| CN-5 | 100 | 50 | 50 | 25 |
| CN-6 | 50 | 50 | 25 | 50 |
| CN-7 | 100 | 50 | 12.5 | 25 |
| CN-8 | 50 | 100 | 50 | 100 |
| CN-9 | 100 | 50 | 100 | 50 |
| CN-10 | 50 | 100 | 100 | 50 |
| CN-11 | 100 | 50 | 25 | 50 |
| CN-12 | 50 | 25 | 25 | 50 |
| CN-13 | - | 100 | 50 | 50 |
| CN-14 | 50 | 100 | 100 | 50 |
| Ciprofloxacin | 3.7 | 3.8 | 3.5 | 3.5 |
Table 7: Antibacterial activity MIC data (μg/ml).
Antimycobacterial activity
According to the MABA report, introduction of sulfonamide moiety increased the antitubercular activity with MIC value at 12.5 μg/ml. Compound-11 had also shown remarkable activity at low MIC value due chlorine atom at its 6th position. Compound-7 has exhibited remarkable activity against standard drug INH (Table 8).
| Compound | MIC (μg/mL) |
|---|---|
| CN-1 | 25 |
| CN-2 | 100 |
| CN-3 | 50 |
| CN-4 | 100 |
| CN-5 | 50 |
| CN-6 | 100 |
| CN-7 | 25 |
| CN-8 | 50 |
| CN-9 | 100 |
| CN-10 | 12.5 |
| CN-11 | 25 |
| CN-12 | 50 |
| CN-13 | 50 |
| CN-14 | 100 |
Table 8: MIC of anti tubercular activity of synthesized compounds.
In silico docking studies
The Mol Dock scores of the fourteen tested compounds range between -93A5 and -13A5 docking studies were performed with antibacterial DNA gyrase B in addition with E.coli in order to understand molecular activity of compounds. According to docking studies carbonyl group of compound 7 interacted with Asn52 of nitrogen group compound 11 had shown interaction with Asn52 of nitrogen and oxygen atom compound 12 also shown three interactions out of which two interact with Ile 84 of oxygen atom and another one with Thr 169 of nitrogen atom. Based on the docking report CN-7 was observed to be a potent compound against E.coli with possible interactions with best docking score. Docking studies with M. tuberculosis DHFRase in addition to E.coli was performed to investigate the activity of main compounds. Owing to docking report of antimycobacterial activity, Ala-7 interacts with oxygen atom of compound-10. All the synthesized compounds exhibited profound activity against microbes. Docking analysis supports antibacterial and antimycobacterial results. Lead compound identification can be better possible with development of cinnoline molecule by optimization of pharmacodynamic and pharmacokinetic properties. In conclusion, the combination of two active rings displayed profound antimicrobial activity.
Antimycobacterial activity
According to the MABA report, introduction of sulfonamide moiety increased the antitubercular activity with MIC value at 12.5 μg/ml. Compound-11 had also shown remarkable activity at low MIC value due chlorine atom at its 6th position. Compound-7 has exhibited remarkable activity against standard drug INH.
CONCLUSION
The present work depicts the significance of synthesized compounds with better activity against bacterial strains and when compared over the standard drug with good percentage of yield. These derivatives had proven to be best potent drug for fighting against microbes. The mol dock scores of the fourteen tested compounds range between -93 and -135. All the synthesized compounds exhibited profound activity against microbes. Docking analysis supports antibacterial and antimycobacterial result. Lead compound identification can be better possible with development of cinnoline molecule by optimization of pharmacodynamic and pharmacokinetic properties.
Novice cinnoline derivatives were synthesized possessing antibacterial activity and antitubercular activity. Titled compounds are afforded by substituting alkyl, halogen groups at 6th and 7th position in basic cinnoline moiety. Antibacterial activity was evaluated by disk plate method for compound (1-14). MIC values of all compounds are between >100 and 12.5 μg/ml. All compounds has shown good activity, among all compound-7 having carboxyl group at 6th position profounded greater activity against gram -ve bacteria when compared with standard drug with MIC 12.5 μg//ml against E.coli. Compound-11 also demonstrated as outstanding compound possessing chlorine atom and nitro group a 6th and 7th position with better MIC value of 25 μg//ml against E.coli. Compound-12 also showed best activity with MIC value of 25 μg//ml and also significant MIC value is tabulated. Compounds are screened against M. tuberculosis H37Rv by MABA method. Surprisingly, all of the compounds reported MIC between >100 and 12.5 μg/ml. Outstanding MIC value was noted for compound-10 with 12.5 μg/ml. In conclusion, the combination of two active rings displayed profound antimicrobial activity.
References
- Wiederhold NP. Infect Drug Resist. 2017;29:249-259.
[Crossref] [Google Scholar] [PubMed]
- Lewgowd W, et al. Int J Pharma Med Chem. 2007;340(2):65-80.
[Crossref] [Google Scholar] [PubMed]
- Fan YL, et al. Euro J Med Chem. 2018;146:554-563.
[Crossref] [Google Scholar] [PubMed]
- Vargas F, et al. J Photochem Photobiol. 2008;92(2):83-90.
[Crossref] [Google Scholar] [PubMed]
- Satyanarayana M, et al. Bioorgan Med Chem. 2008;16(16):7824-7831.
[Crossref] [Google Scholar] [PubMed]
- Wiederhold NP. Infect Drug Resist. 2017;29:249-259.
[Crossref] [Google Scholar] [PubMed]
- Tonk RK, et al. Euro J Med Chem. 2012;57:176-184.
[Crossref] [Google Scholar] [PubMed]
- Ramalingam P, et al. Ind J Heterocyclic Chem. 2006;15(4):359-362.
- Gomtsyan A, et al. J Med Chem. 2005;48(3):744-752.
[Crossref] [Google Scholar] [PubMed]
- Alvarado M, et al. Chem Biodivers. 2006;3(1):106-117.
[Crossref] [Google Scholar] [PubMed]
- Giamarellou H, et al. Antimicrob Agent Chemother. 1975;7(5):688-692.
[Crossref] [Google Scholar] [PubMed]
- Tonk RK, et al. Euro J Med Chem. 2012;57:176-184.
[Crossref] [Google Scholar] [PubMed]
- Bekhit AA. Bollettino Chimico Farmaceutico. 2001;140(4):243-253.
[Google Scholar] [PubMed]
- Barraja P, et al. Bioorgan Med Chem. 1999;7(8):1591-1596.
[Crossref] [Google Scholar] [PubMed]
- Hu YQ, et al. Euro J Med Chem. 2017;133:255-267.
[Crossref] [Google Scholar] [PubMed]