Original Articles: 2024 Vol: 16 Issue: 9
Isolation and Characterization of Multi Drug Resistant Pseudomonas Aeruginosa from Different Hospitals in Iraq
Safae Abdalhamid Bdawi1*, Saleem Obaid Gatia Almawab2, Abdurrahman Ayvaz3
1Department of Natural and Applied Science, University of Erciyes, Kayseri, Turkey
2Department of Medical Laboratory Technologies, University of Alhuda, Al-Najaf, Iraq
3Department of Biology, University of Erciyes, Kayseri, Turkey
- Corresponding Author:
- Safae Abdalhamid Bdawi Department of Natural and Applied Science, University of Erciyes, Kayseri, Turkey
Received: 02-Sep-2024, Manuscript No. JOCPR-24-147147; Editor assigned: 05-Sep-2024, PreQC No. JOCPR-24-147147 (PQ); Reviewed: 19-Sep-2024, QC No. JOCPR-24-147147; Revised: 26-Sep-2024, Manuscript No. JOCPR- 24-147147 (R); Published: 03-Oct-2024, DOI:10.37532/0975-7384.2024.16(9).190
Citation: Bdawi SA et al. 2024. Isolation and Characterization of Multi Drug Resistant Pseudomonas aeruginosa from Different Hospitals in Iraq. J. Chem Pharm. Res. 16:190.
Copyright: © 2024 Bdawi SA. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Abstract
176 swab samples were collected from wound and burn sites at AL-Ramadi Teaching Hospital, Maternity and Children Teaching Hospital in Ramadi, and Karama Hospital in Baghdad during the first quarter of 2019. Out of these, 53 isolates (isolation rate: 30.1%) were identified as Pseudomonas aeruginosa; these included 34 isolates from wound sites and 19 isolates from burns. The isolates were cultured on Blood, MacConkey, and Cetrimide agars and identified using microscopic, phenotypic, and biochemical tests; final confirmation was obtained via API 20E. Antibiotic resistance was assessed using the disc diffusion method against nine antibiotics, showing resistance rates ranging from 22.65% to 82.02%, with tetracycline exhibiting the highest resistance (82.02%) and ceftazidime the least resistant (22.65%). A genetic screening for the blaTEM and pelF genes using RT-PCR revealed that five wound isolates carried the blaTEM gene, while the pelF gene was found primarily in isolates from wound samples. This study highlights significant antibiotic resistance in P. aeruginosa and the usefulness of combining molecular and conventional diagnostic methods to assess bacterial profiles in hospital settings. Minimal inhibitory concentrations were determined using the VITEK 2 system.
Keywords
Pseudomonas aeruginosa; pelF; blaTEM gene; RT-PCR; VITEK 2; API20E
Abbreviations
ICUs: Intensive Care Units; PCR: Polymerase Chain Reaction; API 20E: Analytical Profile Index 20 Enterobacteriacea; MIC: Minimal Inhibitory Concentration; AST: Antibiotic Sensitivity Test; AK: Amikacin; CAZ: Ceftazidime; CFM: Cifixime; CIP: Ciprofloxacin; IPM: Imipenem; NA: Nalidixic Acid; NO: Norfloxacin; TE: Tetracycline; RT-PCR: Real-Time Polymerase Chain Reaction; MDR: Multi Drug Resistant
Introduction
Gram-negative rod-shaped bacteria, Pseudomonas aeruginosa, are an opportunistic pathogen with a range of identified virulence factors that gradually become a severe cause of nosocomial infections. It is one of the most common pathogens in Intensive Care Units (ICUs), Normal and intact skin, as a component of the innate immune system, forms a barrier against infectious agents and prevents pathogenic bacteria from entering the body. However, injuries resulting from burns and other factors impair the physiological function of the immune system, destroying skin protection against infection and increasing the risk of burn patients being infected with nosocomial infectious agents such as Pseudomonas aeruginosa. Considering that 42% to 65% of death rates in burn patients are due to infection, such infections become more concerning [1]. Although this pathogen is not very effective in healthy individuals, it causes high rates of morbidity and mortality, particularly in patients with cystic fibrosis and immunocompromised individuals [2]. It can be found in soil, water, and surfaces, as well as in the natural flora of humans and animals.
Diseases in plants, insects, fish, reptiles, birds, and mammals can be caused by this kind [3]. Because of its wellknown ability to form biofilms, P. aeruginosa can thrive in harsh environments on contaminated sites and in clinical settings [4]. Environment and genetic background are two examples of the factors that influence the formation and behavior of biofilm matrices [5]. Surface-adhered biofilms can be a source of environmental colonization and are well-documented sources of infection in clinical settings [6]. Furthermore, because of the high likelihood of genetic exchange between bacterial species inside the biofilm matrix, biofilms are regarded as hotspots for antibiotic resistance. Furthermore, P. aeruginosa is a significant bacterial species since it can be found on human skin and in moist hospital environments. When the skin is burned or injured, the body's natural defenses are compromised, which creates an environment that is conducive to bacterial growth and reproduction [7]. Numerous burns and wounds are susceptible to infection and complications due to P. aeruginosa.
These bacteria can enter the bloodstream and induce septicemia and bacteremia in immuno compromised leukemia patients. One of the most common nosocomial bacteria is P. aeruginosa and 10%-15% of these pathogens are opportunistic ones that can cause respiratory tract, meningitis, cystic fibrosis, eye, otitis media and pneumonia infections, among other infections [8]. The ability of P. aeruginosa to withstand numerous hospital disinfectants as well as resistance to numerous medications as a result of bacterial mutations has been shown to increase mortality from P. aeruginosa infections [9]. Gram-negative P. aeruginosa possesses virulence factors such as PiliIV, flagellum, exotoxin A (exoA), alkaline protease (aprA), exoenzymes (exoS, exoU, and exoT), the synthesis of elastase and sialidase, and the type III secretion system. Fever and shocks associated with sepsis are caused by these causes [10]. Antibiotic-resistant strains of P. aeruginosa are easily produced. In addition to being widely used and indiscriminate antibiotics, particularly in developing and third-world nations, these bacteria's resistance to several antibodies has been bolstered by the presence of transport plasmids that have generated resistant strains and by the presence of genes that automatically fend off changes in the mechanisms of antibodies [11]. Recent years have seen a rise in the study of P. aeruginosa due to the discovery of numerous genes and other components in the bacteria thanks to the Polymerase Chain Reaction (PCR) Technique. It was discovered that resistance to numerous antibiotics was caused by these genes and mutational factors [10]. Because of the great sensitivity and high accuracy of molecular techniques, they were widely used in the diagnosis of this particular bacterium [12]. Due to the indiscriminate and uncontrolled use of antibiotics, P. aeruginosa antibiotic resistance is a major global issue [13]. In order for bacterial cells to function continuously, gene expression is needed. They constantly modify gene expression by activating or inhibiting particular genes, implicated transcription factors, in response to environmental stresses. Thus, P. aeruginosa was checked for in swaps taken from hospitalized patients in three separate hospitals for this investigation.
Materials and Methods
At the Maternity and Children Teaching Hospital in Ramadi, Al-Ramadi General Teaching Hospital, and Karama Hospital in Baghdad, swabs were taken from patients suffering from burns and surgical wounds. From the start of January to the end of March 2019, a three-month timeframe was used for the sample collection. First, swabs were obtained and grown on MacConkey and blood agar for morphological evaluation. Then, in the event of contamination, P. aeruginosa was found using cetrimide agar. Based on their cultural traits, results from biochemical testing, and appearance under a microscope following gram staining, all of the isolates were identified [14]. After gram staining, the bacterial isolates were seen under a microscope using a bacterial smear to determine the morphology, aggregation, and interaction of the cells with the dye. The BioMeriex (France) API 20E diagnostic kit was used to identify P. aeruginosa oxidase, catalase, and Kligler iron urease-positive isolates. 0.25 ml of saline and bacterial colonies was introduced to each of the biochemical tests. This strip-shaped kit's 20 wells were incubated for 24 hours at 37°C. The number obtained by scoring the groups in the lane was ascertained by comparing it to the P. aeruginosa reference value after reactions in each well were observed.
The VITEK-2 compact systems are utilized to determine the Minimal Inhibitory Concentration (MIC) of antibiotics
P. aeruginosa isolates' Minimum Inhibitory Concentrations (MICs) were ascertained using Antibiotic Sensitivity Test Cards (AST Cards) for the VITEK 2 compact system.We distributed antibiotic sensitivity test cards and evaluated 12 isolates against various drugs. There are multiple concentrations of every antibiotic. As the bacteria grow, the gadget monitors changes in turbidity [15]. Antibiotic Discs: Kirby Bauer created this easy-to-implement, low-cost approach [16]. P. aeruginosa was grown extensively in the medium on paper discs impregnated with varying antibiotic doses. After a full day of incubation, the antibiotics in the discs began to spread towards the agar, and eventually inhibition zones were seen all around the disc. The microorganism's sensitivity to the antibiotic increases with the size of the zone of inhibition surrounding the disc. Next, the findings obtained with various antibiotics were compared, the inhibition zones were measured and recorded in millimeters, and the bacteria's sensitivity to antibiotics was computed as a percentage. The antibiotic discs that were used to isolate microorganisms.
Antibiotic concentrations and icons
The antibiotics of amikacin, ceftazidime, nalidixic acid, tetracycline are 30 μg/ml, ciprofloxacin, gentamicin, imipenem, norfloxacin are 10 μg/ml and Cifixime is 5 μg/ml.
Molecular study
Extraction of genomic DNA: Genomic DNA was extracted from bacterial isolates using the ABIO pure extraction technique. DNA quantification the concentration of extracted DNA was measured using a quantus fluorometer to assess the suitability of the samples for use in subsequent processes. 199 μl of diluted Quantity Flourdye was combined with 1 μl of DNA. DNA concentration readings were found following a 5-minute incubation period at room temperature. Primer pairs intended for the blaTEMand pelF genes were used in real-time PCR analysis of all bacterial isolates.
Genes and primer details:
• Gene: blaTEM
• Annealing Temperature: 55ºC
• Primer Sequences:
Forward: 5'-ATGAGTATTCAACATTTCCG-3'
Reverse: 5'-GACTCTGCAACAAATACGC-3'
• Gene: pelF
• Annealing Temperature: 60ºC
• Primer Sequences:
Forward: 5'-CCTTCTCGGTGTTCTTCATC-3'
Reverse: 5'-GCATTCGCTGGCATAGT-3'
The following protocol was used to carry out this PCR. The primary resolutions employed in this investigation were created in compliance with the company's guidelines. 30 μM of primer was mixed with 300 μM of water and allowed to dissolve to create 100 μM of primer solutions. After that, this solution was diluted 1:10 and kept cold (-20ºC) [17,18].
Statistical analysis
ANOVA was used to analyze the data using SAS's GLM methods (SAS, 2002). Duncan's multiple range tests was used to distinguish the differences between the antibiotic means at a probability value of (P ≤ 0.05). When necessary, we ascertained whether there was a significant difference between the averages of our two samples using Student's test [19].
Results
P. aeruginosa isolation and identification
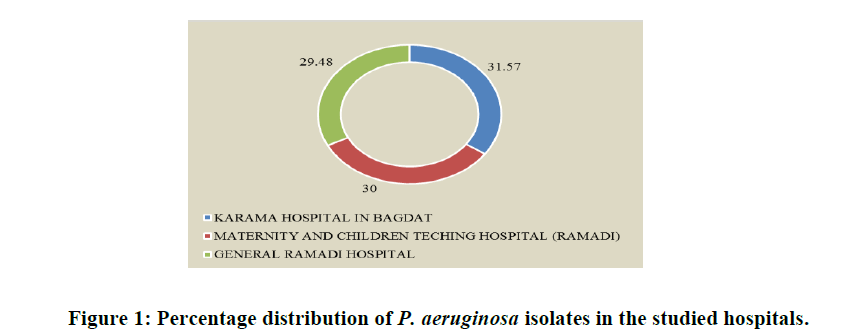
During the months of January through March 2019, P. aeruginosa isolates were tested at the AL Ramadi General Teaching Hospital, the Maternity and Children Teaching Hospital in Ramadi and the Karama Hospital in Baghdad. Patients provided a total of 176 samples, of which 105 (59.65%) had wounds and 71 (40.34%) had burns. Shows that of the 53 P. aeruginosa isolates, 19 (10.79%) were isolated from burns and 34 (19.31%) from wounds. The patients, who ranged in age from 1 to 70 years, included both male and female persons from whose microorganisms were identified in Table 1. The percentage of P. aeruginosa isolates extracted from each institution was discovered to be somewhat similar. For AL-Ramadi General Teaching Hospital, Maternity and Children's Teaching Hospital in Ramadi, and Karama Hospital in Baghdad, the corresponding percentages of positive isolates were 29.48, 30.0, and 31.57%, as shown in Table 2 and Figure 1.
| Source of infection | Number of samples | Number of P. aeruginosa isolates |
|---|---|---|
| Wounds | 105 (59.65%) | 34 (19.31%) |
| Burns | 71 (40.34%) | 19 (10.79%) |
| Total | 176 (100%) | 53 (30.1%) |
Table 1: The distribution of P. aeruginosa isolates by isolation infection.
| Hospital | Total | % of Positive isolates |
|---|---|---|
| AL-Ramadi General Teaching Hospital in Ramadi | 78 | 23 (29.48%) |
| Maternity and children Teaching Hospital in Ramadi | 60 | 18 (30.0%) |
| Karama Hospital in Baghdad | 38 | 12 (31.57%) |
| Total | 176 | 53 (30.11%) |
Table 2: Shows the distribution of isolates of P. aeruginosa by isolation area.
The most frequent sources of P. aeruginosa infections were discovered to be 19 (10.79%) burns and 34 (19.31%) wounds in this investigation of the distribution of isolates by infection site. The goal of this study was to separate P. aeruginosa from specific clinical samples (burns, wounds).
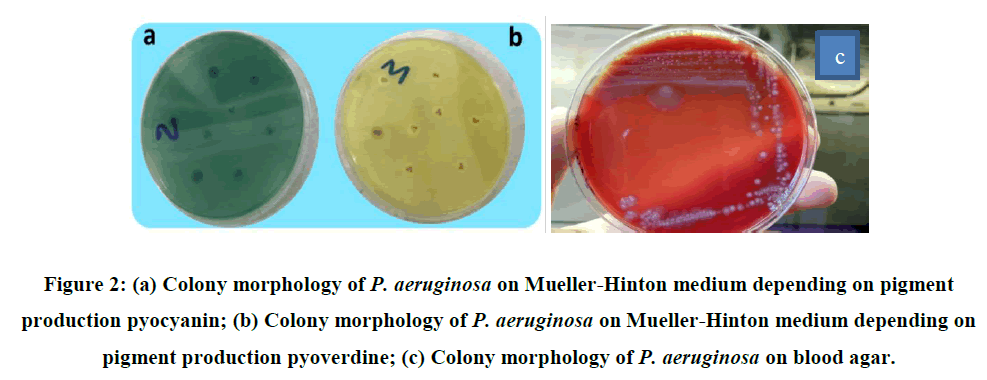
The most frequent sources of P. aeruginosa infections were discovered to be 19 (10.79%) burns and 34 (19.31%) wounds in this investigation of the distribution of isolates by infection site. The goal of this study was to separate P. aeruginosa from specific clinical samples (burns, wounds). During the sampling period, other sources of infection, such as blood, urine, stool, and sputum, were neither included nor excluded. Numerous researchers worldwide have discovered that the recovery rate of P. aeruginosa isolates from the selected clinical samples was higher than expected. Bacterial isolates were morphologically diagnosed using MacConkey agar, which was also utilized to diagnose pale bacterial colonies. Upon examining the morphology of the colonies, it was revealed that the bacteria did not ferment lactose, resulting in uneven colony borders and an off-putting fruity scent. Growing colonies were found to change color in accordance with the pigments that the bacteria produced. Because of the hemolytic enzymes the bacteria produced, when they were cultured on blood agar, β-hemolytic colonies developed. Upon closer inspection, the blood agar colonies were found to be mostly spreading, flat colonies that produced pigment in most cases. It was found that the pigments generated propagated throughout the media, forming colonies that were dark greenish blue in color. Examining the colonies grown on Muller-Hinton agar media, it was found that the bacteria that made pyocyanin produced greenish blue colonies, whereas the bacteria that created pyoverdin produced vivid, yellowish green colonies. It is stated that these pigments are soluble in water (Figure 2)
To ascertain whether the isolates developed in the media during the investigation were P. aeruginosa, a number of physical and chemical tests were conducted. The results are the Gram stain, urease, indole, and methyl red, voges proskauer, growth at 4°C assays yielded negative results, shape cells yielded rods shape results, blood agar yielded β- hemolysis results, cetrimide agar, oxidase, catalase, citrate utilization, growth at 42°C assays yielded positive results, MacConkey agar assays yielded non lactose fermented results, kligler iron agar assays yielded K\K results. Under a microscope, the bacterial preparations made using gram staining revealed that the bacteria were rod-shaped, gramnegative, and did not form spores. It is known that bacteria convert hydrogen peroxide into water and oxygen gas because biochemical studies using bacterial isolates revealed that the bacteria produced positive findings in both oxidase and catalase assays. The bacteria utilized citrate as a carbon source after breaking it down in the positive citrate consumption test. The bacteria in the Kligler iron agar test were found to not create H2S, and the medium's surface was found to be alkaline. P. aeruginosa was identified as the causative agent in the case of medium contamination for some isolates using the centroid agar test. The lactose in MacConkey agar was not fermented by the isolates, as was observed. The isolates could grow at +42ºC and did not develop at -4ºC. The urease, indole, and methyl red assays yielded negative results.
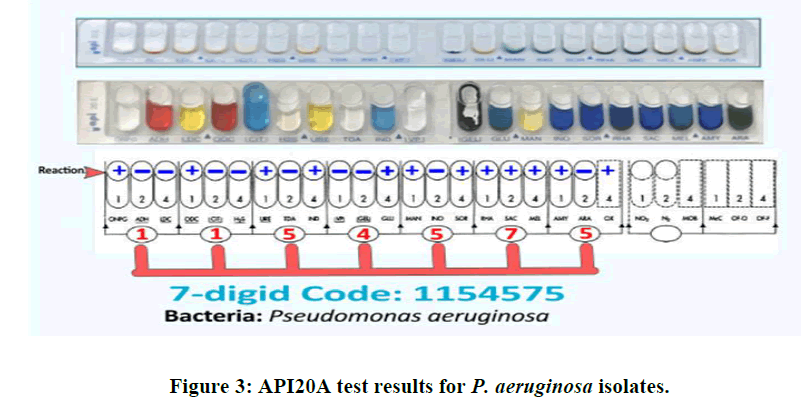
In order to identify isolates resistant to multiple drugs, the biochemical identification test was verified using the API 20E diagnosis system. The API 20E test was run to confirm that all isolates were P. aeruginosa prior to using the VIITEK 2 system to analyze multidrug-resistant bacteria. 12 isolates in total were found to be multidrug resistant following disc diffusion methods. The API 20E test was also used to identify these 12 isolates. The outcomes of the API 20E tests are displayed in (Figure 3). The results of our comparison and scoring system, which was based on API 20E, indicated that these isolates were P. aeruginosa. Following completion of all API 20E system procedures, a seven-digit code was generated by scoring the results in accordance with the Analytic Profile Index (API 20E). The isolates that were examined were identified as P. aeruginosa based on this outcome. Numbers 1, 2 and 4 were used to assess the responses seen in each triple well of the API 20E strip. The scores from each triple system were then compiled to find the seven-digit P. aeruginosa code.
Determination of antibiotics resistance of P. aeruginosa with a disc diffusion method
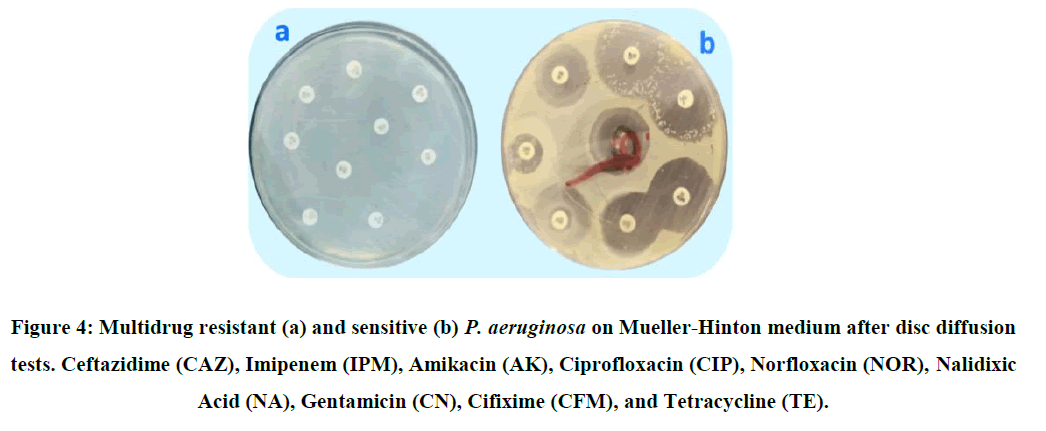
In regular laboratory settings, disc diffusion tests are the most commonly utilized techniques for determining antibiotic sensitivity. Using Hudzicki method, which measures the bacterial isolates' sensitivity or resistance to nine antibiotics based on the diameter of inhibition of the antibiotic discs' surrounding area, the sensitivity of P. aeruginosa isolates to antibiotics was tested (Figure 4) (Table 3) [16]. The study's findings demonstrated that each P. aeruginosa isolate had distinct variations in their resistance to antibiotics. Antibiotic susceptibility testing was performed on 53 isolates that were recognized as P. aeruginosa, as previously mentioned. The results of the sensitivity and resistance tests are displayed in Figure 4 and Table 4. P. aeruginosa isolates were found to be less sensitive to tetracycline (16.98%) and more responsive to ceftazidime (77.35%), as shown in Figure 5.
| Antibiotics | Source | |
|---|---|---|
| Wound | Burn | |
| Ceftazidime | *2.50 ± 0.377bcd | 2.25 ± 0.629bc |
| Imipenem | 2.00 ± 0.462bcd | 2.75 ± 0.478bc |
| Amikacin | 2.87 ± 0.295bcd | 4.00 ± 0bc |
| Ciprofloxacin | 0.668 ± 0.129d | 0.600 ± 0.158c |
| Norfloxacin | 4.12 ± 0.789b | 4.25 ± 1.65bc |
| Nalidixic acid | 2.75 ± 0.619bcd | 4.50 ± 2.02bc |
| Gentamicin | 1.50 ± 0.188cd | 1.25 ± 0.250bc |
| Cifixime | 3.62 ± 0.754bc | 5.25 ± 1.70b |
| Tetracycline | 10.50 ± 1.50a | 10.00 ± 2.58a |
| P-value | 0.0001 | 0.0029 |
Note: * Means ± standard error. Means within each row followed by the same latter are not significantly different (P>0.05).
Table 3: Mean Minimum Inhibitory Concentrations (MIC, µg/ml) of nine different antibiotics in the VITEK 2 system against P. aeruginosa.
| Antibiotics | Total number of P. aeruginosa isolates | Percent sensitivity | Percent resistance |
|---|---|---|---|
| Ceftazidime | 53 | 77.35 | 22.65 |
| Imipenem | 53 | 75.47 | 24.53 |
| Amikacin | 53 | 56.6 | 43.4 |
| Ciprofloxacin | 53 | 43.39 | 56.61 |
| Norfloxacin | 53 | 43.39 | 56.61 |
| Nalidixic acid | 53 | 35.84 | 64.16 |
| Gentamicin | 53 | 28.3 | 71.7 |
| Cifixime | 53 | 22.64 | 77.36 |
| Tetracycline | 53 | 16.98 | 83.02 |
Table 4: Shows how sensitive the P. aeruginosa isolates were to various antibiotics using disc diffusion techniques.
The isolates' determined antibiotic resistance, derived from several sources. Upon analyzing the antibiotic susceptibility of the isolates derived from the burn and wound samples, it was ascertained that the most efficacious antibiotics were Ceftazidime and Imipenem, which the isolates from both the burn and wound samples had comparable sensitivity to. When P. aeruginosa isolates from various sources were evaluated for their sensitivity to Amikacin antibiotics, it was shown that the isolates from the burn had a higher sensitivity than the isolates from worn samples (t=1.95; df=51, P<0.05). P. aeruginosa isolates were discovered to have sensitivity levels below 50% against the antibiotics Ciprofloxacin, Norfloxacin, Nalidixic Acid, Gentamicin, Cifixime, and Tetracycline, as shown in Table 5.
| Antibiotics | Number of isolates | Percent sensitivity (%) | Percent resistance (%) | |||
|---|---|---|---|---|---|---|
| Wounds | Burns | Wounds | Burns | Wounds | Burns | |
| Ceftazidime | 34 | 19 | 76.47 | 78.95 | 23.53 | 21.05 |
| Imipenem | 34 | 19 | 73.53 | 78.95 | 26.47 | 21.05 |
| Amikacin | 34 | 19 | 50 | 68.42 | 50 | 31.58 |
| Ciprofloxacin | 34 | 19 | 44.12 | 42.11 | 55.88 | 57.89 |
| Norfloxacin | 34 | 19 | 41.18 | 47.37 | 58.82 | 52.63 |
| Nalidixic acid | 34 | 19 | 35.29 | 36.84 | 64.71 | 63.16 |
| Gentamicin | 34 | 19 | 29.41 | 26.32 | 70.59 | 73.68 |
| Cifixime | 34 | 19 | 20.59 | 26.32 | 79.41 | 73.68 |
| Tetracycline | 34 | 19 | 14.71 | 21.05 | 85.29 | 78.95 |
Table 5: Comparison of the sensitivity P. aeruginosa isolated from different sources.
These isolates also showed notable resistance to these medications. These isolates were obtained from both wound and burn samples. Tetracycline was shown to be highly resistant among the tested antibiotics, and only 14.71% and 21.05%, respectively, of the isolates from burns and wounds were found to be responsive to these antibiotics. The disk diffusion method was used to determine the isolates' sensitivity to antibiotics. The mean values obtained by measuring each isolate's inhibition zone for each drug are provided in 4.15. Thus, zone diameter was computed at a similar rate for isolates from both wound and burn samples, with the biggest inhibitory zone seen in isolates treated with ceftazidime. Tetracycline and Cifixime showed the lowest inhibition zones, and it was discovered that the isolates from both sources were resistant to these two medications, as shown in Table 6.
| Antibiotics | Source | |
|---|---|---|
| Wound | Burn | |
| Ceftazidime | *12.73 ± 1.75a | 18.47 ± 2.85a |
| Imipenem | 10.82 ± 1.51a | 16.73 ± 2.59a |
| Amikacin | 6.50 ± 1.41b | 11.31 ± 2.12b |
| Ciprofloxacin | 4.70 ± 1.16bc | 6.89 ± 2.08bc |
| Norfloxacin | 3.94 ± 1.00bcd | 6.26 ± 1.74bcd |
| Nalidixic acid | 2.58 ± 0.760cd | 4.05 ± 1.35cd |
| Gentamicin | 2.20 ± 0.646cd | 2.31 ± 1.08cd |
| Cifixime | 0.735 ± 0.281d | 2.31 ± 0.924cd |
| Tetracycline | 0.676 ± 0.347d | 1.00 ± 0.535d |
| P-value | 0.0001 | 0.0001 |
Note: * Means ± Standard Error.
Table 6: The mean zone of inhibition obtained using a different concentration of nine antibiotics against P. aeruginosa.
The means in each row that are followed by the same later do not differ substantially (P>0.05) medicines that combat P. aeruginosa. The disk diffusion method was used to screen the isolates known to be P. aeruginosa in order to identify those that were multi-drug resistant and where they came from. Shows that 12 isolates out of 53 were found to be multidrug resistant, 8 of these isolates were taken from burn samples, and 4 of the isolates were taken from wounds. It was shown that 23.52% of the isolates recovered from samples of wounds were MDR; for burns, the corresponding ratio was 21.05% in Table 7.
| Sources | Total isolates | No. of. MDR isolates | Percentage MDR isolates |
|---|---|---|---|
|
Wounds |
34 |
8 |
23.52 |
|
Burns |
19 |
4 |
21.05 |
|
Total |
53 |
12 |
22.64 |
Table 7: Proportion of P. aeruginosa isolates resistant to multiple drugs from various sources.
Extraction of DNA from P. aeruginosa isolates and its concentration and purity
Utilizing a quantus fluorometer, the concentration of DNA was found. Twelve bacterial isolates with burns and wounds that were resistant to all antibiotics had their DNA extracted. The amount of DNA in the bacterial isolates was assessed and the results are 20 ng/μl in isolate SE1, 25 ng/μl in isolate SE2, 15 ng/μl in isolate SE3, 22 ng/μl in isolate SE4, 25 ng/μl in isolate SE5, 20 ng/μl in isolate SE6, 21 ng/μl in isolate SE7, 20 ng/μl in isolate SE8, 24 ng/μl in isolate SE9, 26 ng/μl in isolate SE10, 20 ng/μl in isolate SE11, 21 ng/μl in isolate SE12. The concentration varied between 15 ng/μl in isolate SE3 and 26 ng/μl in isolate SE10.
Detection of specific resistance genes in P. aeruginosa by RT_PCR
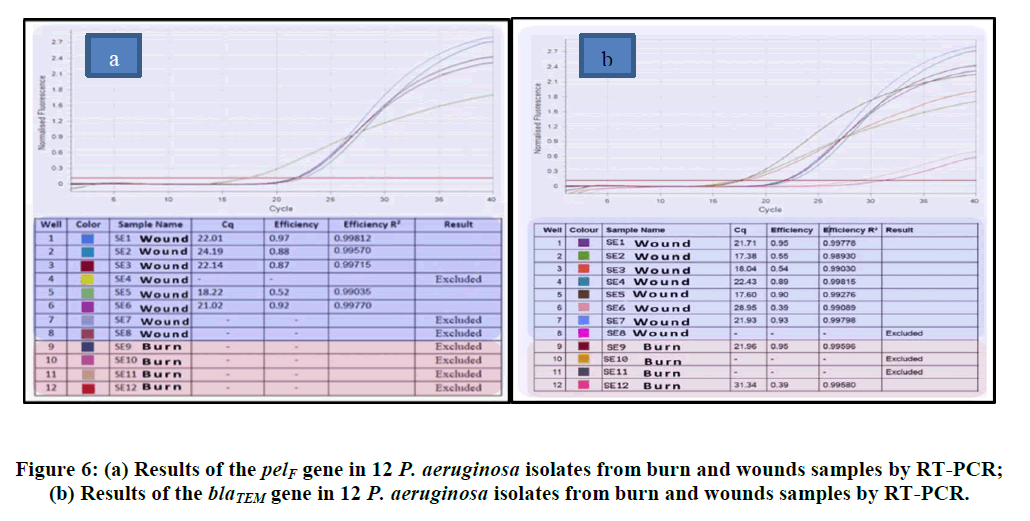
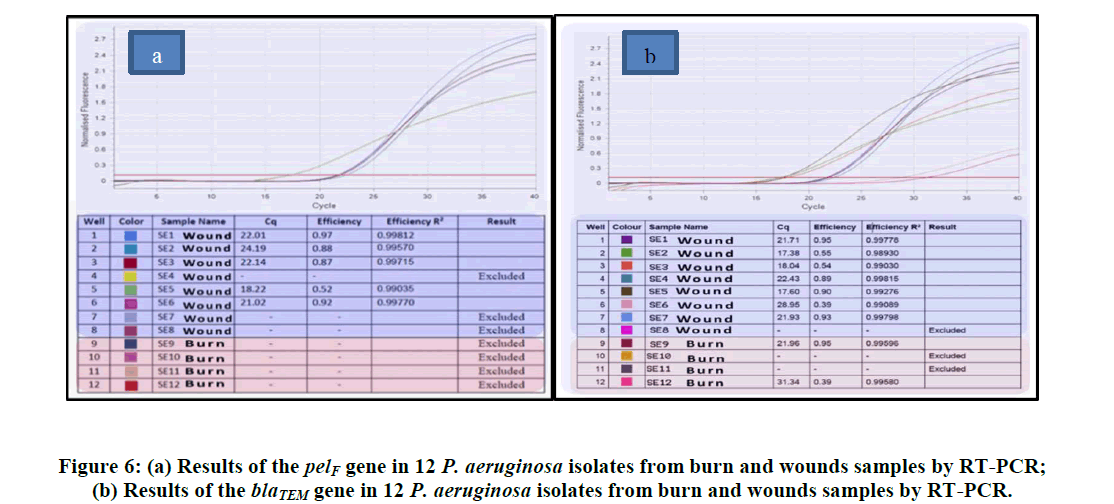
The twelve P. aeruginosa isolates that were classified as Multi Drug Resistant (MDR) were examined using the realtime PCR method to see if they possessed the blaTEM, pelF, and resistance genes. The blaTEM gene was in charge of multidrug resistance, while the pelF gene was in charge of biofilm production. Of these twelve isolates, nine were taken from the wound and four from samples of burns. RT-PCR research revealed that the pelF gene was present in 7 (87.5%) of the isolates obtained from wounds and 2 (50%) of burn samples. Relative to isolates carrying pelF, fewer isolates had the blaTEM gene. It was found that none of the burn samples contained the blaTEMgene; just four of the wound samples did. The Figures provides the relevant statistical data along with isolates that yield positive results (Figure 6).
Discussion
Due to the indiscriminate and uncontrolled use of antibiotics, P. aeruginosa antibiotic resistance is a serious global issue [20]. Continuous activity in bacterial cells necessitates the expression of certain genes that are engaged in transcription factors in response to environmental stimuli. Numerous studies have shown that despite a variety of preventative measures, including as isolation methods and antibiotic de-escalation therapy, the growth in multi-drug resistance nosocomial bacteria continues to pose a concern to hospitalized patients. Current research has concentrated on the impact of P. aeruginosa strains resistant to multiple drugs on burns and wounds because it has been demonstrated that these bacteria have a significant role in the morbidity and death of immuno compromised people [21]. As a result, P. aeruginosa was checked in samples taken from patients with burn and wound issues who were admitted to three separate hospitals for this study. Using the VITEK 2 system as a comparison, we examined the bacteria's resistance to several drugs using the traditional disc diffusion approach. By using RT PCR, resistant gene profiles (pelF and blaTEM) of these isolates were also found. Under the supervision of a medical specialist, samples were taken from patients at the AL Ramadi general teaching hospital, the Maternity and Children Teaching Hospital in Ramadi, and the Karama Hospital in Baghdad who were suffering from burns and wounds. As indicated in table (3.2) and picture (3.2), the ratio of 53 P. aeruginosa isolates to 30.11% was found after the final diagnosis. In our study's human clinical samples, P. aeruginosa was found in all cases at a rate of 30.1%. Similar incidence rates from Georgia (31.5%), Turkey (16.4%), Brazil (37.3%), India (29.6%), Norway and Sweden (25.8 to 45.9%), and Georgia (31.5%) have all been reported (Ranjan KP, Ranjan N, Bansal SK, 2010) [22].
In England, Wales, and Northern Ireland, the total incidence of P. aeruginosa was 6.4 cases/100,000 populations [23]. People 75 years of age and older reported higher rates of infections, particularly in male patients (52.0 cases/100,000 population) as opposed to 19.7 cases/100,000 in female patients (the same age group) 358 patients with P. aeruginosa bacteremia were reported to have been hospitalized in medical wards (133; 37%), ICUs (103; 29%), surgical wards (97; 27%), and neonatology units (25); of these patients, 45 (12%) had HIV infection and 28 (8%) had hematologic malignancies. Analogous studies on hospital infections in Iran have been conducted [24]. The results of this study are in line with several regional and international investigations, such as the one conducted by Alkhalsy, which demonstrated that P. aeruginosa bacteria are the most often isolated bacteria from burns and wounds [25]. Examining the P. aeruginosa isolation research, it was discovered that the number of isolates collected varied depending on a number of parameters, including the geographical makeup of the studied area, the source of isolation, the quantity of samples, improper use of antibiotics, and many others. Whether the patient was taking medications that prevent bacteria from growing or that are resistant to it, which also played a major role in the spread, as well as the level of cleanliness and the kind of disinfectants used in hospitals 21.2% of this was done in the United States [26]. The method of sterilizing wounds or burns and the quantity of sterilizations performed all contributed significantly to the emergence of resistance to these bacteria, which is one of the reasons for the variation in isolation rates. Other factors include the location, number, and source of samples [27].
Pale color colonies were seen and lactose fermentation was not observed when we examined the morphological identification of isolates on the McConkey agar [28]. A similar condition was reported in the investigations [29]. It is known that the colonies grown on the blood agar media are beta hemolytic, and the medium turns greenish blue due to the hemolytic enzymes produced. It has been demonstrated that bacterial colonies on Cetrimide agar are green. Because of the pyocyanin on Mueller-Hinton media, certain P. aeruginosa isolates generated greenish blue colonies,whereas others developed yellowish colonies because of the pyoverdine pigment. Studies have reported the formation of water-soluble pigment [30]. The isolates were identified under a microscope as gram-negative, rod-shaped, nonspore- forming cells. These results were found to be compatible with the findings [28]. All isolates underwent oxidase and catalase tests as part of the biochemical testing, and the results of each experiment demonstrated that the bacteria convert hydrogen peroxide into water and oxygen gas.
Our isolates produced alkaline findings on the Kligler iron agar test and metabolize citrate. The isolates were able to grow at +42?C and did not develop at -4?C. The urease, indole, and methyl red assays yielded negative results. The results aligned with the research conducted [31]. The isolates were identified as P. aeruginosa by applying the API 20E test, which yielded a 7-digit code, 1154575, for each isolate across all applications. Studies have revealed that these bacteria are responsible for many infections, burns, and wounds, particularly in hospitalized patients in intensive care units. P. aeruginosa is one of the most significant Pseudomonas species due to its high illness incidence. It is the primary cause of ICU infection, particularly in patients receiving care in burn units. It is an opportunistic pathogen [32]. Research indicates that burns connected to sepsis account for 75% of fatalities, particularly in developing nations and burnt lobes [33]. The existence of P. aeruginosa strains may be the result of these bacteria's opportunistic behavior, which allows them to exploit a widespread or localized weakness in either or both of the body's immunological or mechanical defenses [34]. The hands of hospital employees, the instruments they use, and the environment of the hospital can all harbor bacteria. These bacterial infections are caused by adhesion factors and antibiotic resistance, as previously mentioned. By measuring the diameter of the inhibitory zone surrounding the disc, the Kirby Bauer method was used to assess the sensitivity or resistance of P. aeruginosa isolates to nine different antibiotics. It was found in these trials that the isolates were most sensitive to ceftazidime, that the susceptibilities of the antibiotics varied, and that the isolates were most resistant to tetracycline. These two antibiotics were shown to be the most effective when tested, as the isolates' susceptibilities to imipenem and ceftazidime were similar. Similar rates of 77.35% were achieved in our investigation. In another study on Pseudomonas aeruginosa, it was stated that the bacteria show 81% susceptibility to this Ceftazidime [35,36]. When an antibiotic is discovered to be effective, its use gradually rises, which encourages the emergence of resistance [37,38]. Comparable outcomes against these drugs were discovered by a few other researchers [39].
According to Al-Arnaudi's findings, 81% of the 75 P. aeruginosa isolates were found to be resistant to Ceftazidime [35]. A ratio of 86% was found in the 250 P. aeruginosa isolates that found to be resistant to this antibiotic [40]. The antibacterial potency of bacteria belonging to the Monobactams category, such as Imipenem and Meropenem, as well as the findings, were found to be 73.3% in a study conducted on 100 P. aeruginosa isolates that were isolated from burns and wounds in Isfahan [39]. The percentage of the antibiotic Amikacin resistant to P. aeruginosa was 20 percent. The current study's findings resembled those in certain ways [41]. 25% of P. aeruginosa infections in hospitals have been isolated. The study's findings disagreed [42]. Tetracycline resistance was found in 83.02% of cases. This is in line with the results, who reported that 100% of P. aeruginosa isolated from various infections was present [43].
The antagonist's inability to reach the cell's inhibitory concentration is the cause of this resistance. This is the outcome of the plasmids' programmed processes, which either decrease the antibody's absorption or make it easier for it to leave the cell. P. aeruginosa bacteria produce the enzymes known as "Cephalosporinase Penicillinase," which attack the beta-alkaloids in the nucleus of penicillins and cephalosporins, rendering them ineffective and generating extended-spectrum beta-lactamaes, which is one of the main causes of the bacteria's resistance to beta-lactam antibodies and experiencing a shift in the permeability of the outer membrane as a result of the closure of outer membrane apertures [44,45]. The current study's findings demonstrated that P. aeruginosa bacteria that are resistant to the medications gentamicin and amikacin are members of the aminoglycoside antibiotic class. It was noted by the researcher Morita et al., that the current study's results regarding the proportion of anti-resistance Amikacin at 70%, along with the resistance ratio of 74% Gentamicin, were noteworthy [46]. The study conducted by Khorsheed et al., on 28 isolates of P. aeruginosa bacteria revealed that the local isolates exhibited resistance to the aminoglycosides group, and that the amikacin resistance ratio was 64% [47]. The development of bacteria is the cause of antibiotic resistance to this class of drugs. Enzymes like N-acetyl transferase and phosphor transferase have been altered by P. aeruginosa. These enzymes' genes are based on chromosomes or plasmids [39]. Additionally, to resistance brought on by chromosomal abnormalities or changes in membrane permeability [48]. The isolates of P. aeruginosa in this investigation, however, demonstrated resistance to Quinolones, such as Ciprofloxacin, Norfloxacin, and Nalidixic acid in contrast, the findings reported 23 isolates of P. aeruginosa, Ciprofloxacin (74.62%), and the study [48,49]. They discovered that 187 P. aeruginosa isolates were resistant to 71.1% of ciprofloxacin. It is despised that 34.0% of people were resistant to norfloxacin. P. aeruginosa's resistance to quinolones is caused by either a mutation in the DNA enzyme or an inhibition of DNA synthesis via DNA gyrase inhibition. Emphasized that, in order to ascertain MIC values and improve the precision of antibiotic classification, a diffusion approach ought to be used in addition to VITEK 2. Twelve P. aeruginosa isolates that were found to be multidrug resistant by various means were tested for blaTEM and pelF gene presence using RT-PCR. Bacteria that form biofilms become naturally resistant to antimicrobial treatments and can cause chronic illnesses. Thus, a deeper comprehension of the genetic and molecular processes underlying biofilm development can lead to crucial treatment and prophylactic antibiotic resistance treatments. According to PCR results, the isolates from wound samples, SE1 through SE7, were positive for the PelF primer, whereas SE8 did not have this gene. Because the SE7 isolate produced more mRNA and had higher levels of expression, it was able to build more biofilm. However, isolate SE6 had lower expression of this gene and was unable to form as much biofilm. Two of the four isolates from the burn samples were found to possess this gene and it expressed itself low in SE12 and highly in SE9. Once more, the blaTEMgene was screened by RT PCR from these 12 resistant isolates. The results showed that while none of the isolates from the burn samples had this gene, five isolates from wound samples-SE1, SE2, SE3, SE5, and SE6-did. This gene was predominantly expressed in the SE1 isolate and at least in the SE5 isolate, according to the RT-PCR findings we looked at. This finding implied that isolate SE1 would be more resistant to antibiotics containing beta lactam than other isolates.
Conclusions
• The extensive and unregulated use of antibiotics has led to a global worry regarding antibiotic resistance in P. aeruginosa.
• Specific genes involved in transcription factors are activated or inhibited by bacterial cells in response to environmental stressors.
• Hospitalized patients are still at risk from multi-drug resistant nosocomial bacteria, such as P. aeruginosa, despite measures such isolation procedures and antibiotic de-escalation therapy.
• Recent research has concentrated on the effects of multi-drug resistant P. aeruginosa strains on burns and wounds because these infections are a major cause of morbidity and mortality in people with weakened immune systems.
• Patients with burn and wound issues from three separate hospitals provided samples for this investigation.
• Using the traditional Disc Diffusion approach, the resistance of P. aeruginosa bacteria to different antibiotics was evaluated and contrasted with the VITEK 2 system.
• Using RT-PCR, the study also found that these P. aeruginosa isolates had resistance gene profiles (pelF and blaTEM).
• Out of the patients sampled, 53 isolates of P. aeruginosa were found, representing 30.11% of the total incidence.
Recommendations
• A suggestion to lessen the indiscriminate and uncontrolled use of antibiotics in an effort to address the bacterial resistance issue.
• It is advised to keep an eye on P. aeruginosa gene expression in order to learn how the bacteria react to environmental stressors by either activating or inhibiting particular genes linked to genetic transcription.
• A suggestion to improve hospital infection control protocols in order to reduce the transmission of bacteria resistant to multiple drugs and safeguard hospitalized patients.
• The recommendation to carry out additional research to comprehend the effects of P. aeruginosa strains that are resistant to drugs on burns and wounds, as these bacteria are a leading cause of morbidity and death in people with impaired immune systems.
Declarations
Availability of data and materials
Data and materials are available on request.
Authors' Contribution
S. BDAWI carried out experiments, analyzed the data, and wrote the manuscript. S. ALMAWA and A. AYVAZ analyzed and proofread the first draft. All authors approved the final version of the manuscript.
Acknowledgement
I want to convey my heartfelt gratitude to my mam for her tremendous support and assistance in completing my project. I would also like to thank our Principal, and all of my friends, for providing me with this wonderful opportunity to work on this project. The completion of the project would not have been possible without their help and insights.
References
- Chastre J, Fagon JY. Am J Respir Crit Care Med. 2002;165(7):867-903.
[Crossref] [Google Scholar] [PubMed]
- Zavascki AP, Barth AL, Fernandes JF, et al. Crit Care. 2006;10:1-7.
[Crossref] [Google Scholar] [PubMed]
- Holban AM, Chifiriuc MC, Cotar AI, et al. Rom Biotechnol Lett. 2013;18(6):8843-8854.
- Singh R, Bishnoi NR, Kirrolia A. Bioresour Technol. 2013;138:222-234.
[Crossref] [Google Scholar] [PubMed]
- Chayabutra C, Ju LK. Appl Environ Microbiol. 2000;66(2):493-498.
[Crossref] [Google Scholar] [PubMed]
- Wei Q, Ma LZ. Int J Mol Sci. 2013;14(10):20983-21005.
[Crossref] [Google Scholar] [PubMed]
- Kaiser SJ, Mutters NT, DeRosa A, et al. Eur J Clin Microbiol Infect Dis. 2017;36:243-253.
[Crossref] [Google Scholar] [PubMed]
- Malhotra S, Limoli DH, English AE, et al. mBio. 2018;9(2):10-128.
[Crossref] [Google Scholar] [PubMed]
- Sastry AS, Bhat S. JP Medical Ltd; 2018.
- Al-Daraghi WA, Abdullah ZH. Al-Nahrain J Sci. 2013;16(2):167-172.
- Mitiku M, Ali S, Kibru G. Am J Biomed Life Sci. 2014;12(3):40-45.
- Finnan S, Morrissey JP, O'gara F, et al. J Clin Microbiol. 2004;42(12):5783-5792.
[Crossref] [Google Scholar] [PubMed]
- Mavrodi DV, Bonsall RF, Delaney SM, et al. J Bacteriol. 2001;183(21):6454-6465.
[Crossref] [Google Scholar] [PubMed]
- Baron SMP, Finegold EJ. Mosby Company. Missouri. 2007.
- Pincus DH. Biomerieux. 2006;2006:1-32.
- Hudzicki. J Am Soc Microbiol. 2009 Dec 8;15(1):1-23.
- Peymani A, Naserpour-Farivar T, Zare E, et al. J Prev Med Hyg. 2017;58(2):E155.
[Crossref] [Google Scholar] [PubMed]
- Ninyio NN, Gidado HB, Yahaya MO, et al. Afr J Biotechnol. 2017;16(12):585-593.
- Duncan DB. biometrics. 1955;11(1):1-42.
- Mitiku M, Ali S, Kibru G. Afr J Biotechnol & Life Sci. 2014;12(3):40-45.
- Dantas SR, Moretti-Branchini ML. Infect Control Hosp Epidemiol. 2003;24(5):351-355.
[Crossref] [Google Scholar] [PubMed]
- Ranjan KP, Ranjan N, Bansal SK, et al. J Lab Physicians. 2010;2(02):74-77.
[Crossref] [Google Scholar] [PubMed]
- Burjanadze I, Kurtsikashvili G, Tsereteli D, et al. Int J Infect Control. 2007;3(2).
- Japoni A, Alborzi A, Kalani M, et al. Burns. 2006;32(3):343-347.
[Crossref] [Google Scholar] [PubMed]
- Alkhalsy AHA. Master Thesis, Faculty of Basic Education, Mustansiriya University. 2014.
- Dharan S, Mourouga P, Copin P, et al. J Hosp Infect. 1999;42(2):113-117.
[Crossref] [Google Scholar] [PubMed]
- ALmosafer HC. Master Thesis, Faculty of Science, Mustansiriya University, pages. 2007;108.
- Forbes BA, Sahm DF, Weissfeld AS. Mosby; 2007.
- Selim S, El Kholy I, Hagagy N, et al. Biotechnol Biotechnol Equip. 2015;29(1):152-156.
[Crossref] [Google Scholar] [PubMed]
- Sudhakar T, Karpagam S, Premkumar J. J Chem Pharm Res. 2015;7(3):921-924.
- M. Tadesse,. Alem A. EPHTI .Gondar University. 2006.
- Patil P, Joshi S, Bharadwaj R. Int J Healthcare Biomed Res. 2015;3(3):106-112.
- Al-Aali KY. Arch Clin Microbiol. 2016;7(2):1-9.
- Brooks GF, Carroll KC, Butel JS, et al. Sultan Qaboos Univ Med J. 2007;7(3):273.
- Al-Arnaudi AFM. Coll. Educ. Pure Sci. Ibn al-Haytham, Univ. 2015;123.
- Ruh E, Gazi U, Güvenir M, et al. Turk Hij Den Biyol Derg. 2016;73(4):333-344.
- Gallini A, Degris E, Desplas M, et al. J Antimicrob Chemother. 2010;65(12):2650-2657.
[Crossref] [Google Scholar] [PubMed]
- Vernaz N, Huttner B, Muscionico D, et al. J Antimicrob Chemother. 2011;66(4):928-935.
[Crossref] [Google Scholar] [PubMed]
- Vaez H, Safaei HG, Faghri J. Burns Trauma. 2017;5.
[Crossref] [Google Scholar] [PubMed]
- Al-Shwaikh RM. elife. 2016.
- Shahid M, Malik A. Indian J Med Res. 2005;122(4):324.
[Google Scholar] [PubMed]
- Mandal MD, Mandal S. Asian Pac J Trop Biomed. 2011;1(2):154-160.
[Crossref] [Google Scholar] [PubMed]
- Jalil MB, Abdul-Hussein ZR, Al-Hmudi HA. Int J Dev Res. 2017;7:11.
- Morita Y, Tomida J, Kawamura Y. Front Microbiol. 2012;3:408.
[Crossref] [Google Scholar] [PubMed]
- Khorsheed MB. Kirkuk J Sci. 2017;12(1).
- Ochoa SA, Lopez-Montiel F, Escalona G, et al. J Bol Med Hosp Infant 2013;70(2):133-144.
- Al-Saray ZA.
- Yusuf E, Van Herendael B, Verbrugghe W, et al. Ann Intensive Care. 2017;7:1-7.
[Crossref] [Google Scholar] [PubMed]
- Ekrem K, Rokan DK. Sky J Microbiol Res 2014;2(2):13-17.